Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Protein Baby Pictures
An NMR study enables analysis of a protein while it's still busy being born on the ribosome
by Stu Borman
October 22, 2007
| A version of this story appeared in
Volume 85, Issue 43
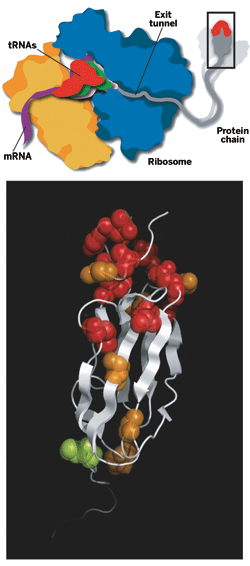
WHEN A BABY is born, one of the first things that one wants to do is take pictures of it. But for baby proteins newly biosynthesized on ribosomes, obtaining structural pictures has proved extraordinarily difficult. After a nine-year effort, a European group has now achieved that long-sought goal by structurally analyzing and characterizing a nascent protein chain while it was still being created.
The study shows that a baby protein can fold at least partially into its native three-dimensional form while it's still attached to its parent ribosome. Ribosomes are the cellular machines charged with creating proteins. There was considerable evidence that newborn proteins did fold on the ribosome, with or without assistance from large protein complexes called molecular chaperones. But a detailed snapshot of the phenomenon had never before been captured.
The ability to analyze proteins on the ribosome could open a new area of research on the relationship between protein biosynthesis and folding. It could also lead to a more detailed understanding of the mechanism by which molecular chaperones promote protein folding and to strategies to prevent protein misfolding, which can cause neurodegenerative diseases.
The work was carried out by Paola Fucini, now a professor of structural biology at the University of Frankfurt, in Germany; chemistry professor Christopher M. Dobson of the University of Cambridge; John Christodoulou, now a lecturer in structural biology at the University of London; and coworkers, including Cambridge postdoc Shang-Te Danny Hsu (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.0704664104). They obtained the structure by using an ultrafast nuclear magnetic resonance (NMR) spectroscopy technique to analyze a nascent protein assembled from isotope-labeled amino acids.
PERFORMING SUCH an analysis has heretofore been "an impossible experiment," Dobson says. The ribosome is a huge biomolecular assembly containing over 7,500 amino acids in more than 50 proteins, and its NMR spectra are normally much too complex to permit the analysis of a small protein chain attached to it. The researchers solved this problem by exploiting the ability of NMR to "see" localized regions that move independently within large biomolecules. A nascent protein chain on the ribosome has sufficiently rapid and extensive motion to give NMR lines as narrow as those of small proteins in solution.
The researchers found that a nascent two-domain immunoglobulin protein folds at its N-terminal end while its C-terminal end is still stuck in the ribosome exit tunnel, the birth canal for baby proteins. The N-terminus of a protein always emerges first during biosynthesis. The analysis shows that the protein's N-terminal area contacts the ribosome while it folds, suggesting that the ribosome may assist protein folding, as predicted earlier by other groups.
F. Ulrich Hartl, a chaperone expert at Max Planck Institute of Biochemistry, Martinsried, Germany, says the study "represents a major breakthrough in that it demonstrates the feasibility of NMR structural analysis of nascent polypeptide chains and extends what can be done by NMR."
Ribosome specialist Peter B. Moore, a professor of chemistry at Yale University, points out that a very early antecedent of the new work was a study by David Zipser, now professor emeritus of cognitive science at the University of California, San Diego, showing that β-galactosidase folds sufficiently on the ribosome during its synthesis to display enzymatic activity (J. Mol. Biol. 1963, 7, 739). Given these observations, Moore says, protein folding on the ribosome "is not a surprise."
Nevertheless, Moore adds, "Who would have thought that you could make a large enough sample of pure nascent chain complex to do NMR on? Ten years ago, who would have believed, even if the preparative problems could be solved, that useful NMR spectra would result? What this paper does report is a tour de force in both preparative biochemistry and NMR."
Biomolecular NMR specialist Ad Bax of the National Institute of Diabetes & Digestive & Kidney Diseases, Bethesda, Md., says the work represents a major advance in the NMR study of very large complexes. He notes that to make the study feasible the group "used the latest in NMR hardware—700- and 900-MHz machines, equipped with cryogenically cooled receivers for the highest achievable sensitivity—combined with recent advances in NMR pulse sequence development."
Chemistry professor Lewis E. Kay of the University of Toronto, who also specializes in biomolecular NMR, says: "Emerging NMR technologies developed in the past few years are now making it possible to begin to see nascent protein structure on the ribosome and to ask questions about the dynamics of that structure that have been elusive until now. This paper is a very important first step" toward that goal.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter