Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Helping Biology Catch Up
Phosphoproteomics needs to progress from cataloguing sites to understanding their biology
by Celia Henry Arnaud
March 10, 2008
| A version of this story appeared in
Volume 86, Issue 10
ONE WAY THAT CELLS communicate is by adding or removing phosphate groups at specific locations along proteins. Mass spectrometry is a powerful tool for identifying such phosphorylation sites. Large-scale mass spectrometry-based experiments have created vast catalogs of phosphorylation sites. As is the fashion of the day when it comes to data- and molecule-intensive biological science, this activity has become associated with one of the many "omics" fields—in this case, phosphoproteomics.
Extracting the most value from phosphoproteomics, experts say, will require moving beyond catalogs. The power of mass spectrometry for identifying phosphorylation sites on proteins has outpaced the ability of biology to determine the function of those sites, says Michael B. Yaffe, a biochemist and bioengineer at Massachusetts Institute of Technology.
"The thing that's typically done is to grab a sample, grind it up, throw it in the mass spectrometer, and map as many sites as you possibly can without a lot of thought to the biology that went into" creating those phosphorylation sites, says Forest White, associate professor of biological engineering at MIT.
Current phosphoproteomics methods are not without their challenges. Matthias Mann, a mass spectrometrist at the Max Planck Institute for Biochemistry in Martinsried, Germany, notes that identifying phosphorylation sites using mass spectrometry is more difficult than simple protein identification with this tool. With protein identification, multiple peptides can point to the same protein, improving the accuracy of the identification. In phosphoproteomics, however, each peptide stands alone, and the location of the phosphorylation site within the peptide and the overall protein needs to be pinpointed. "All this means that it is technically more demanding," Mann says.
In addition, "phosphorylation is often substoichiometric," says John Yates, a professor of chemical physiology at the Scripps Research Institute in LaJolla, Calif. "A protein may not go from 0% phosphorylated to 100% phosphorylated. It may go from 10% phosphorylated to 40% phosphorylated."
As such, proteomics leaders advocate quantitative approaches that focus on identifying phosphorylation changes under different conditions, such as the presence or absence of growth factors or other signaling molecules. "The more complicated the biological system you're looking at is, the more important it is to have a contrast," Yates says.
Mann and his coworkers have undertaken one such large-scale study to quantify cellular responses to stimulation with epidermal growth factor (EGF). They identified roughly 6,600 phosphorylation sites in cellular proteins, and only about 600 sites changed as a result of EGF stimulation (Cell 2006, 127, 635). With the latter sites, "you're in good shape, because you know that this is the protein, it phosphorylates here, and that phosphorylation site changes when you put the growth factor on the cell," Mann says. "Almost by definition, this site must have a role in growth-factor signaling."
White worries that even such contrast experiments, which he also does, are just quantitative cataloging. "It's like a catalog with prices, which beats a catalog with no prices," he quips.
But such quantitative catalogs may not provide the answers people seek. "This doesn't necessarily give us what those phosphorylation sites are doing biologically. It just gives the idea that the ones that aren't changing probably aren't implicated in the biology," White says.
Figuring out the function of a particular phosphorylation site is made even more difficult by the adaptability of biological systems. "When you knock out a gene, it changes everything else around it. The same is true for phosphorylation sites," White says. Converting an amino acid that can be phosphorylated (serine, threonine, or tyrosine) to one that can't be phosphorylated affects more than that individual site. "The cells are going to adapt, and you have to deal with that," White says.
To confront these confounding issues, White's group quantifies phenotypic changes in addition to quantifying the phosphorylation changes. For example, in a study of the effects of HER2 (a type of epidermal growth factor receptor known to be involved with cancer) overexpression, White quantified cancer-cell proliferation and migration in addition to phosphorylation changes under a variety of conditions. White and Douglas Lauffenburger, who is also at MIT, analyzed the data to find the phosphorylation sites that correlated most strongly with the phenotype of cancer-cell proliferation and migration.
Using a combination of computational analysis and phenotypic characterization with quantitative cataloguing, White and Lauffenburger prioritize phosphorylation sites for more in-depth biological experiments. "You can think of it as target ranking," White says.
An additional challenge for determining biological function is that phosphoproteins are present at such low abundance that large numbers of cells are needed to perform the analysis. Ideally, phosphoproteomics would be performed at the single-cell or even subcellular level.
"We're looking at averages across a bunch of cells," Yates says. If you want to determine absolutely what a protein with a particular pattern of phosphorylation is doing, he says, "you have to be down at the single-cell level."
BIOINFORMATICS IS KEY to navigating the large amounts of data. Yaffe and Anthony J. Pawson of Mount Sinai Hospital in Toronto developed a program called NetworKIN that combines the sequence motifs that are phosphorylated by kinases (the enzymes that add phosphate to proteins) with mass spectrometry data to build pathways that connect kinases and phosphorylated proteins (Cell 2007, 129, 1415).
"The big gap has always been trying to identify the substrates of a kinase that account for the behavior we see," Yaffe says. The combination of mass spectrometry data and bioinformatics allows Yaffe to ask the questions: "Of all these sites that have been phosphorylated and cataloged, which ones were generated by our kinase of interest? If the kinase we're interested in really phosphorylated that substrate, would it explain any of the biology we see?"
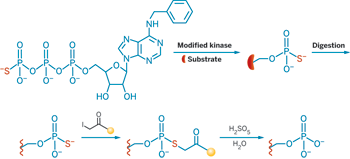
Kevan M. Shokat, a Howard Hughes Medical Institute investigator at the University of California, San Francisco, and UC Berkeley, has recently described a method to draw those connections between kinases and their substrates (Proc. Natl. Acad. Sci. USA 2008, 105, 1442).
In Shokat's method, a kinase is engineered to accept an adenosine triphosphate analog that tags the protein substrate with a phosphothioate group instead of the usual phosphate group. He uses this phosphothioate tag to capture the kinase's substrate.
Using this method, Shokat's team identified substrates for the kinase Cdk1-cyclin B, which is involved in mitosis. The researchers found many candidate substrates, including ones that were already known to be Cdk substrates.
Mass spectrometry has been the tool of choice for finding phosphorylation sites, and it will continue to play a role in the future. But other methods may someday prove to be more useful for studying the biological function of known sites. "It's hard to imagine monitoring the signaling network across many perturbations in many different samples by mass spectrometry because it's just not high-throughput enough," White says. "Protein microarrays are probably going to be the wave of the future."




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter