Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Choose Your Own Analytical Range
Biosensors: Mixtures of receptors with different affinities enable expansion of dynamic range
by Celia Henry Arnaud
January 9, 2012
| A version of this story appeared in
Volume 90, Issue 2

By combining sets of DNA receptors with different binding affinities, a research team has created biosensors that detect specific DNA sequences over customized concentration ranges (J. Am. Chem. Soc., DOI: 10.1021/ja209850j). The approach extends concentration range without impairing specificity, a trade-off that has plagued past biosensor range-improvement efforts.
Kevin W. Plaxco, a chemistry professor at the University of California, Santa Barbara, whose group carried out the study, notes that conventional single-site DNA receptors have a fixed 81-fold dynamic range. Dynamic range is the difference between lowest and highest concentrations that can be detected reliably, defined by the researchers as representing 10% and 90% receptor occupancies, respectively. Nature deals with this restriction by combining multiple receptors to achieve the dynamic range that natural sensors require for various functions, Plaxco says.
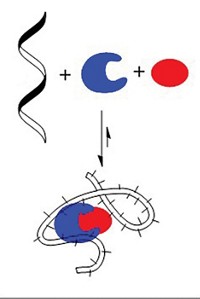
Plaxco and his coworkers decided to do their own mixing and matching of receptors to manipulate biosensor dynamic ranges. They generated families of molecular beacons—stem-loop DNAs that fluoresce upon binding other specific DNA sequences—by making modifications that don’t perturb the beacons’ DNA-binding sites.
Plaxco’s team mixed various combinations of the molecular beacons to create biosensors with different dynamic-range profiles. A 59/41 mixture of a pair of receptors with 100-fold affinity difference results in a sensor with 8,100-fold useful concentration range.
Pairs of receptors with larger affinity differences produce biosensors with significant deviations from linearity in the intermediate portion of their response curves. To achieve wider but useful dynamic ranges, the researchers combined larger numbers of receptor variants. A mixture of four different receptors—two with 10,000-fold affinity difference and two with a smaller affinity difference—results in a sensor that detects an approximately 900,000-fold range of concentrations.
Plaxco and coworkers also modulated how the biosensors respond to concentration changes in several ways. In one case they constructed sensors with linear responses at the lower and upper ends of the concentration range but with an almost flat response at intermediate concentrations. In other cases, they designed sensors with narrower dynamic range but improved sensitivity to small concentration differences.
An advantage of the new devices over previous extended-range biosensors is that modifying receptors at sites remote from their DNA-binding sites expands range without impairing DNA sequence specificity, comments Sergey N. Krylov, a chemistry professor at York University, in Toronto, who has also worked to extend biosensor dynamic range. “I look forward to seeing if similar modifications can be done for aptamers [synthetic DNA sequences] that bind protein targets,” he says.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter