Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
A Quick Snapshot of Protein Aggregation
Mass Spectrometry: Electrospray ionization provides a fast analysis of the dynamic process
by Leigh Krietsch Boerner
January 4, 2012
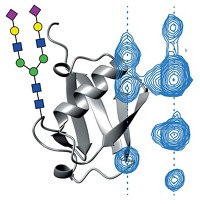
Researchers have developed a high-resolution technique to analyze the size of protein aggregates as they form (Anal. Chem., DOI: 10.1021/ac203017x). They hope the method, which uses electrospray ionization mass spectrometry, will help scientists to study the molecular mechanisms behind diseases such as Alzheimer’s and to control the quality of protein-based drugs.
While protein aggregation is often associated with disease, protein clumps also cause dilemmas for the pharmaceutical industry. Protein-based drugs can aggregate during production or storage, which can bury the active parts of the proteins, rendering the drugs partially or fully inactive.
Existing techniques to follow and characterize protein aggregation have significant limitations, says Igor Kaltashov, professor of chemistry at the University of Massachusetts, Amherst. Methods such as analytical ultracentrifugation and size exclusion chromatography can provide only low-resolution data. The poor resolution, he says, makes it difficult to distinguish between a tetramer and a pentamer, for example. Kaltashov also points out that size exclusion chromatography can take tens of minutes, a delay that means researchers can miss important events as the proteins aggregate.
“There are few methods to see the aggregation process as it goes forward,” says Kaltashov. “We’re good at seeing the end result, but what happens in between is very difficult to track.”
So Kaltashov and his team turned to electrospray ionization mass spectrometry because the technique can produce rapid, high-resolution data. To watch the proteins aggregate, they first had to heat their samples to trigger the proteins to unfold and start to clump up. They tested their method using human antithrombin III, a glycoprotein important to blood coagulation. The researchers followed the aggregation process and found that the mass spectrometry data matched data collected by size-exclusion chromatography.
Monitoring protein aggregation “is difficult by any means of analytical techniques,” says Albert Heck of Utrecht University, in the Netherlands. But he says the mass spectrometry method has a clear advantage, because it easily separates each mass of the different-sized aggregates. He thinks that the method will find most use in the biopharmaceutical industry.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter