Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
The Technical Side: Sequencing Methods Used In Clinical Genetic Testing
by Celia Henry Arnaud
July 15, 2013
| A version of this story appeared in
Volume 91, Issue 28
Clinical DNA sequencing started with Sanger sequencing. That’s the method used in the Human Genome Project, and it’s still used clinically for tests that require sequencing only a small portion of the total genome, such as single-gene tests.
COVER STORY
The Technical Side: Sequencing Methods Used In Clinical Genetic Testing
In the Sanger method, DNA polymerase replicates a single-stranded DNA template using a mixture of four normal deoxynucleotides (one for each type of DNA base) and four fluorescently labeled dideoxynucleotides. Dideoxynucleotides are added sequence-specifically but randomly during the replication process, and each time that happens the reaction stops, resulting in a mixture of DNA strands of various lengths. Those reaction products are separated by size. Because each dideoxynucleotide is labeled with a fluorescent dye that emits light at a different wavelength, the fluorescence identifies the base at that location. The sequence is read out as a ladder.
Sanger sequencing is cost-effective for small-scale sequencing, such as for individual genes. Next-generation sequencing technologies are much more cost-effective for analyses requiring the sequencing of larger stretches of the genome, such as multiple genes, the entire protein-coding portion of the genome (the exome), or the whole genome.
All next-gen sequencing methods start with the construction of a library of small DNA fragments from the region to be sequenced. Those fragments are then sequenced. A number of next-gen sequencing technologies are available, but the ones that dominate the clinical market are Illumina’s MiSeq and HiSeq and Life Technologies’ Ion Torrent.
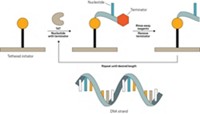
In the MiSeq and HiSeq systems, clusters of sequencing templates, each with a different fragment of the target DNA sequence, are bound to the surface of a flow cell. The reagents needed for DNA synthesis are added to the flow cell. All four fluorescently labeled nucleotides are added to the sequencing reaction simultaneously, but only the one complementary to the next base in a template is successfully added. The nucleotide contains a reversible terminator, which temporarily stops the reaction, and a fluorescent label that identifies the base. Fluorescence imaging of the flow cell reveals which base was added to different templates. Then the terminator is removed, and the next base in the sequence is analyzed. Millions of reactions occur simultaneously.
Life Technologies’ Ion Torrent sequencing systems are similarly based on high-throughput, massively parallel sequencing by synthesis, but they use natural nucleotides instead of fluorescent ones. The synthesis reactions occur on a semiconductor chip that has micromachined wells on top of an ion-sensitive layer. The four different DNA nucleotides are added sequentially. Every time one of them is incorporated into the growing DNA strand, a hydrogen ion is released. Any well in which a base is added will therefore experience a pH change that is detected by the chip as a voltage change. If the base is not complementary, it is not added, and there’s no voltage change to detect. By keeping track of which bases were added in which wells, the sequence at those locations can be identified.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter