Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Starving Bacteria To Find Antibiotics
Drug Discovery: Nutrient-restriction technique finds metabolic blockers
by Stu Borman
October 21, 2013
| A version of this story appeared in
Volume 91, Issue 42
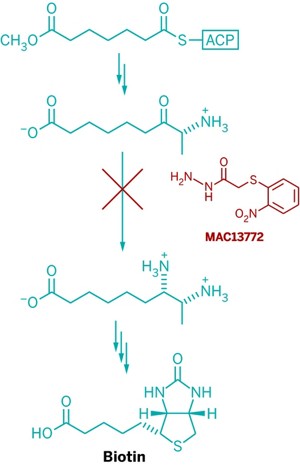
Bacteria are clever at side-stepping antibiotics. They’re so good at developing resistance to medicines that some infections can no longer be treated effectively, making the search for new antibiotics a top priority.
More agents that block bacterial metabolism could be valuable additions to the antibiotic arsenal, but they’re rare and difficult to identify. A new approach for screening compound collections could make them easier to find (Nat. Chem. Biol. 2013, DOI: 10.1038/nchembio.1361).
Compounds are usually tested for antibacterial activity against robust bacteria grown in nutrient-rich cell culture media. In the new approach, Soumaya Zlitni, Lauren F. Ferruccio, and Eric D. Brown of McMaster University, in Hamilton, Ontario, grow bacteria on a restricted diet, a condition known to increase their vulnerability to agents that affect their metabolic pathways. Brown notes this is a realistic situation for screening potential antibiotics because bodies of infected patients are also nutrient-limited environments for bacterial growth.
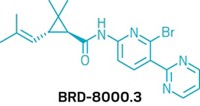
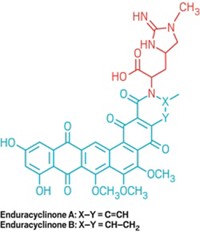
Brown’s group used the technique to discover three synthetic compounds that interfere with bacterial metabolic pathways—glycine metabolism, p-aminobenzoic acid biosynthesis, and biotin biosynthesis. The researchers determined the three compounds’ mechanisms of antibacterial action by testing affected bacteria with specific metabolites to identify the step in each pathway that the compounds block.
Two current antibiotics also block bacterial metabolism. But up to now there hasn’t been a systematic way to search for such agents, and the new technique could help fill that gap. “We think the approach may have potential as a commercial platform,” Brown says.
The technique “is a useful probe for antibiotics,” comments pathogen therapeutics expert Deborah Hung of Massachusetts General Hospital and the Broad Institute, in Boston. “I’m hesitant to say this is going to change antibiotic discovery. But it highlights an important issue—the question of which targets are essential in the body during bacterial infection, where exact microenvironments may vary or are difficult to define. And it’s a valuable approach for understanding the mechanisms by which newly identified small molecules block bacterial metabolism.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter