Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
NMR-EPR Combo Yields RNA-Protein Structure
Spectroscopy: Approach illuminates how translation activator works
by Jyllian Kemsley
May 19, 2014
| A version of this story appeared in
Volume 92, Issue 20

A new technique that combines nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) spectroscopy has enabled researchers to determine the structure of a noncoding RNA bound to its target proteins. The structure helps explain how the RNA activates translation in bacteria by soaking up repressor proteins like a sponge.
Many important macromolecules, particularly those involving RNA, are structurally organized but also very floppy. Such extremely flexible macromolecules can be difficult to structurally characterize. The ability of the new combined NMR-EPR approach to resolve solution structures of RNA and RNA-protein complexes makes it a “very powerful” tool, says Hashim Al-Hashimi, a biochemistry professor at Duke University School of Medicine who was not involved in the new work.
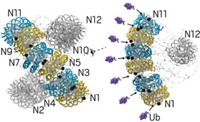
In developing the new technique, a team at ETH Zurich led by former graduate student Olivier Duss and molecular biology professor Frédéric H.-T. Allain investigated the noncoding RNA RsmZ and its interactions with the protein RsmE (Nat. Commun. 2014, DOI: 10.1038/ncomms4669 ).
RsmZ activates translation by sequestering RsmE, which otherwise inhibits translation by binding to messenger RNA. The researchers found that RsmZ contains eight protein-binding sites and can bind five RsmE protein dimers in a sequential manner.
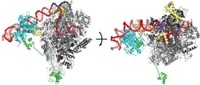
To get the RsmZ-RsmE structure, they first broke the RNA into chunks that each bind one protein dimer. They used NMR to determine the structures of the smaller pieces. Then, they created full-length RNA with added spin labels that allowed them to use EPR to get long-range distances of the full RNA-protein complex. The distances constrain how the smaller pieces fit together into the bigger complex. The team ultimately made 21 different spin-labeled constructs to characterize two different conformers of the complex.
The structures provide insight into how RsmZ soaks up RsmE to permit translation. RNA-protein binding is cooperative—one dimer induces changes to the RNA structure to promote binding of subsequent dimers. Dimer binding also covers nuclease-binding sites, protecting the RNA from degradation.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter