Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Designer Ribosome Works In Live Cells
Synthetic Biology: Researchers engineer entire protein-making complex for first time
by Stu Borman
July 30, 2015
| A version of this story appeared in
Volume 93, Issue 31

Researchers have artificially engineered a complete ribosome—the cell-based machine that translates mRNA into proteins—in the laboratory.
The ability to engineer the ribosome not only could help scientists understand how the protein-maker works, but it also could endow the ribosome with new functions. For drug discovery and basic research, engineered ribosomes could create nonnatural proteins or even nonprotein polymers that would be difficult or impossible for native ribosomes to make.
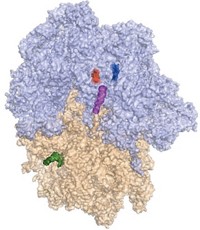
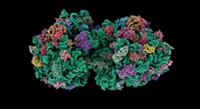
Alexander Mankin of the University of Illinois, Chicago; Michael C. Jewett of Northwestern University; and colleagues designed, engineered, and characterized the artificial ribosome, which they call Ribo-T (Nature 2015, DOI: 10.1038/nature14862).
“It’s an impressive body of work that will enable the directed or random evolution of ribosomes with modifications that allow synthetic amino acids to be more efficiently incorporated, or even to expand the genetic code,” comments ribosome assembly specialist Katrin Karbstein of Scripps Research Institute Florida.
Ribosomes have two independent parts, small and large subunits, which come together in cells to form a complete structure when protein translation is needed. Small ribosomal subunits that make a specific protein have been prepared by genetic engineering before, but not the entire ribosome.
Mankin, Jewett, and coworkers made Ribo-T by engineering ribosomal RNAs, the main components of the small and large subunits, into a single hybrid gene that included two short polyadenine RNA linkers to connect the RNAs. Then the team introduced the modified gene into bacteria. The bacterial cells transcribed the gene into tethered ribosomal RNAs, which then joined with ribosomal proteins made by the cells to form the complete ribosome.
Ribo-T can replace all of a bacterium’s natural ribosomes, can express all native proteins in the bacterial genome, and works nearly half as fast as native ribosomes—fast enough to sustain normal cell growth and proliferation. The researchers also demonstrated that it could be engineered to make a protein with an amino acid sequence native ribosomes can’t handle.
Scientists previously believed that, for ribosomes to work properly, their two subunits had to be independent and had to come together only when needed. But Ribo-T, with its linked subunits, seems to disprove that.
Ribo-T “is a long-sought platform for exploring the mechanism of protein synthesis,” comments ribosome mechanism expert Jamie H. D. Cate of the University of California, Berkeley.
Mankin, Jewett, and coworkers want to use Ribo-T to explore poorly understood ribosome functions and make novel protein-based agents for drug discovery and other applications. “Our new protein-making factory holds promise to expand the genetic code in a unique and transformative way,” Jewett says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter