Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Designed enzyme rescues live bacteria
Protein designed from scratch is efficient enough to perform essential biological function in cells
by Stu Borman
January 18, 2018
| A version of this story appeared in
Volume 96, Issue 4

It’s hard to get proteins designed from scratch, called de novo proteins, to catalyze reactions nearly as efficiently as native enzymes do. Although de novo-designed proteins have shown off their catalytic prowess in lab glassware, getting them to catalyze reactions at levels that can affect the biology of living organisms hasn’t been easy.
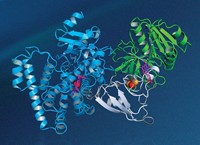
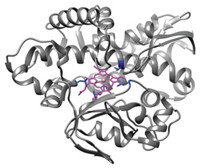
Michael H. Hecht and coworkers at Princeton University have done so by designing a de novo protein that sustains living bacteria genetically modified to require its catalytic activity (Nat. Chem. Biol. 2018, DOI: 10.1038/nchembio.2550). The work could lead to other de novo enzymes that perform useful biological roles in cells.
F. Akif Tezcan’s group at the University of California, San Diego, previously mutated a natural redox protein to perform β-lactamase activity in bacteria. And Hecht’s group showed earlier that a de novo protein could sustain living bacteria lacking an essential gene, but most likely worked by nonenzymatic means. The new study shows that de novo proteins can perform essential catalytic roles in cells.
Hecht’s group started with a de novo four-helix protein called Syn-IF. When expressed in Escherichia coli mutated to remove ferric enterobactin esterase, Syn-IF allowed the microbes to survive under low-iron conditions. The natural esterase’s activity also helps bacteria thrive in low-iron environments. But Syn-IF showed no detectable enzymatic activity. The researchers next mutated it and screened for mutants that could sustain low-iron bacteria better than Syn-IF did. The result was Syn-F4, which catalyzed the same reaction as the natural esterase.
The new study “is significant and exciting,” says Ivan V. Korendovych of Syracuse University, not only because Syn-F4 works in bacteria but also because it has a simple four-helix structure, raising the possibility of designing comparably simple proteins that catalyze other reactions in live organisms.
Syn-F4 “not only replaced the function of a naturally evolved enzyme but, remarkably, used a completely different sequence, structure, and mechanism to do so,” adds Burckhard Seelig of the University of Minnesota. “The ability to create non-natural enzymes that function in a living organism is crucial to realize the hopes of synthetic biology.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter