Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Genomics
Large genomics effort yields antiparasite targets
Bioinformatics approach finds already-approved drugs that might hit the targets
by Megha Satyanarayana
November 8, 2018
| A version of this story appeared in
Volume 96, Issue 45

In underdeveloped and developing nations, more than 1 billion people suffer from parasitic infections. Because existing treatments are lacking, many of those affected must deal with the consequences of the infections, including chronic pain, disability, and lost productivity, throughout their lives. These consequences make parasitic infections a major global health and economic concern.
The need for new drugs is critical, says Peter Hotez, dean of the National School of Tropical Medicine at Baylor College of Medicine, but the motivation to develop them isn’t always there. And the lack of involvement from pharmaceutical companies has meant that it’s hard to find funding for clinical trials for potential new drugs, he says.
An international group of researchers is now asking whether it’s possible to repurpose already-approved drugs to kill tapeworms, hookworms, and the other tiny burrowing creatures that set our immune systems ablaze. To answer the question, they’ve turned to genomes—a staggering 81 to be exact.
The group, called the International Helminth Genomes Consortium, has released early findings of a genetic expedition to find new targets for drug discovery in dozens of species of flat- and roundworms that infect humans and livestock. The targets that it has pulled out include proteases, enzymes that tamp down the immune system, and proteins that allow worms to digest their hosts as the worms move through them. The team has also created a means to predict which existing drug molecules could hit these newfound targets (Nat. Genet. 2018 DOI: 10.1038/s41588-018-0262-1).
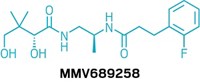
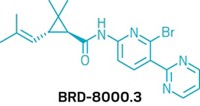
After sequencing 45 new genomes of different worms, Avril Coghlan, a bioinformatician at the Wellcome Sanger Institute, and colleagues combined them with existing genomes for 36 other parasites. They ran the data through software that helped them identify large gene families present in parasitic worms that might not exist in nonparasitic worms. They compared those resulting possible parasite targets with known targets of existing drugs in a small-molecule database called ChEMBL. They then built a list of about 1,200 high-probability matches between drugs with known activity and their parasite targets. That list included molecules like zolmitriptan, which treats migraines, and odanacatib, which is an investigational compound that affects bone resorption.
Hotez says the study is important because worms are the most common human pathogen, and having the genetics from so many species will aid drug development. Coghlan and colleagues made the genomes available at WormBase ParaSite..
Although finding known compounds that can hit parasite targets is the best bet for a fast-track approval for new treatments, it’s also intellectually intriguing to find targets in the data for which there is no known molecular hit, says Makedonka Mitreva, one of the lead researchers in the consortium. “They are very attractive but more challenging to start working with,” she says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter