Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Synthetic Biology
Platform controls proteoglycan architecture
Proteoglycan editing tool will help elucidate how different parts of the molecule contribute to function
by Celia Henry Arnaud
May 26, 2022
| A version of this story appeared in
Volume 100, Issue 19

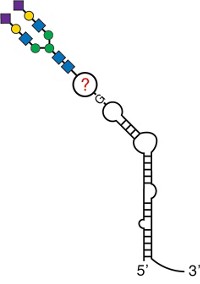
Proteoglycans—molecules consisting of a core protein with multiple sugar chains—are so complex that researchers usually study the protein and sugar components separately, which makes it difficult to understand their overall structure and function. Mia L. Huang and coworkers at Scripps Research in California now report a platform for chemically editing proteoglycans. They have already used the platform to study a proteoglycan involved in the spread of cancer cells (Nat. Chem. Biol. 2022, DOI: 10.1038/s41589-022-01023-5).
The platform lets them precisely control the location and composition of the sugar chains—known as glycosaminoglycans, or GAGs—on proteoglycans. GAGs are long sugars with repeating disaccharide units made of a glucuronic acid and an amino sugar. The disaccharide composition and the glycosidic linkages dictate how the GAGs are classified, the most common being heparan sulfates and chondroitin sulfates.
The researchers’ method involves engineering both protein and GAG components. They used unnatural amino acids with alkyne handles to define the attachment points for the sugar chains on the protein cores. They used azide-linked monosaccharides in order to hijack the biological machinery to make GAGs. The resulting GAGs added to the protein core via a click-chemistry reaction between the alkynes and azides.
For example, the team used the platform to make synthetic versions of various syndecans, a widely studied family of proteoglycans. “We didn’t want to veer too far away from what nature was trying to tell us,” Huang says. “Our initial goal was to be able to ask questions like, ‘Why does nature make these molecules in the way that it does?’”
The researchers used the platform to study the roles of each component of a proteoglycan in the spread of cancer cells. They found that the core protein alone was not sufficient to promote cell spreading—the GAGs were also needed. They also found that the proteoglycan needed to be anchored to the cell membrane rather than in a soluble form outside the cell.
“This method brings us an important step closer to recapitulating the natural system. The ability to mimic full-size proteoglycans and precisely engineer their structures is impressive,” says Linda Hsieh-Wilson, an expert on glycobiology at the California Institute of Technology. “Such tools will enable systematic investigations into different proteoglycan architectures.” In addition, she says, “the work presents another important technology for remodeling glycans on cell surfaces.”
The researchers focused on the portion of proteoglycans on the surface of cells; they didn’t include the portions that are embedded in the cell membrane or inside the cell. The inability to look at those portions of the proteoglycans is a limitation of the method; Huang and her team are working on ways to overcome that limitation.
Huang plans to use the platform to study neurexin, which is involved in the formation of synapses and has only recently been shown to be a proteoglycan. “To me, that discovery was just like, shoot, we don’t know everything yet about all possible proteoglycans in the cell,” she says. She intends to combine her new platform with other methods that identify molecules that are interacting with one another to figure out how neurexin’s interactions change as a function of glycosylation patterns.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter