Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Synthesis
Making the ‘crystalline sponge’ more user-friendly
Multiwell microplate protocol boosts yields of quality crystals for X-ray analysis and cuts down on experiment time
by Bethany Halford
November 28, 2016
| A version of this story appeared in
Volume 94, Issue 47
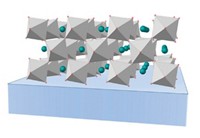
In 2013, when University of Tokyo’s Makoto Fujita reported a way to determine a complex molecule’s X-ray crystal structure without having to actually crystallize the molecule in question, the technique was hailed as a breakthrough for structural determination. But the technique, known as the “crystalline sponge” method because guest molecules soak into a metal-organic framework crystal where they orient themselves in a way that their structure can be determined via X-ray analysis, has yet to be adopted as a routine characterization tool. The technique, as originally reported, takes a significant amount of trial-and-error. Only about 5% of the crystals prepared are suitable for soaking, and it can take up to 16 days to create a single guest-soaked crystalline sponge. Seeking a way to shorten and improve this process, University of Illinois, Chicago, researchers Bernard D. Santarsiero, Neal P. Mankad, and Greyson W. Waldhart made some tweaks to existing protocols (Org. Lett. 2016, DOI: 10.1021/acs.orglett.6b03119). Rather than grow crystalline sponges in a batch process as the original protocol indicates, the UIC researchers used multiwell microplates and layered a drop of ZnI2 in methanol with a drop of the sponge ligand 2,4,6-tris(4-pyridyl)-1,3,5-triazene in nitrobenzene. This step produces crystals, 90% of which are suitable sponges, in only 10 hours. Because it takes seven days for this step under the original protocol, this improvement dramatically cuts down on experiment time.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter