Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Antibiotics
Potential antibiotic treats tuberculosis
Adding lipid chains makes tunicamycin analogs that treat tuberculosis in mice
by Celia Henry Arnaud
November 12, 2018
| A version of this story appeared in
Volume 96, Issue 45

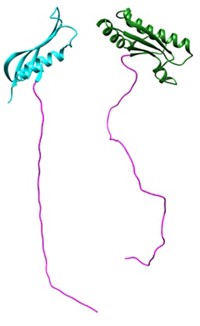
Streptomyces bacteria produce tunicamycin, a compound that shows promise as an antibiotic because it can inhibit cell-wall synthesis in other bacteria. Unfortunately, tunicamycin isn’t suitable for clinical use because it inhibits the human enzyme DPAGT1, which plays an important role in protein production. To find safe alternatives, researchers synthesized tunicamycin derivatives and studied the structure and function of DPAGT1. A team led by Benjamin G. Davis at the University of Oxford added lipid chains of varying lengths to two positions on the core tunicamycin scaffold. To understand the functional significance of these changes, Elisabeth P. Carpenter’s team at Oxford solved structures of the human enzyme alone as well as bound to its natural substrate and tunicamycin (Cell 2018, DOI: 10.1016/j.cell.2018.10.037). The human protein has a narrow tunnel that can bind only one lipid chain on tunicamycin, while the bacterial target has a wide groove that can accommodate two. The most potent analogs, called TUN-8,8 (shown, lipid chains in red) and TUN-9,9, had lipid chains eight and nine carbons long, respectively. In mouse studies, the analogs safely and effectively treated tuberculosis.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter