Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein Breakdown Recounted
Chemistry Nobelists describe research path that led to prize-winning ubiquitin findings
by STU BORMAN, C&EN WASHINGTON
November 1, 2004
| A version of this story appeared in
Volume 82, Issue 44
It was the road not taken. In the late 1970s, hot areas of biochemistry research included such fields as protein synthesis. But graduate student (now professor) Aaron Ciechanover and professor Avram Hershko of Technio--Israel Institute of Technology, in Haifa, were interested in the opposite of protein synthesis: regulated protein breakdown, the process by which cells get rid of proteins that are problematic or have outlived their usefulness. Enzymologist Irwin A. Rose of Fox Chase Cancer Center, Philadelphia (now at the University of California, Irvine), later joined them in their quest.

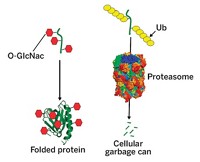
And the rest, as they say, is history. Ciechanover, Hershko, and Rose will now share the nearly $1.4 million 2004 Nobel Prize in Chemistry for having discovered ubiquitin-mediated protein degradation. In this process, chains of the peptide ubiquitin mark unwanted proteins and direct them to a large protein complex, the proteasome, which then breaks them down into peptide fragments that can be reused.
"I'm really overwhelmed" about sharing the Nobel Prize, Ciechanover told C&EN the week of the announcement (C&EN, Oct. 11, page 10). "It's a happy thing, which I have not begun to digest yet"--no pun intended.
Regulated protein degradation is now known to play an important role in cell division, DNA repair, protein synthesis, and the immune system. And disease processes such as inflammation, muscle atrophy, and cancer result from dysregulation of the degradative process. A drug that inhibits the proteasome--Millennium Pharmaceuticals' multiple myeloma agent, Velcade--is the first protein degradation inhibitory agent to have been approved, and others are currently being investigated.
Research on the ubiquitin system can be traced back about a half century. According to Rose, "The whole thing started with the realization that ATP [adenosine triphosphate] was required for protein breakdown"--an observation made in liver slices in 1953 by biochemist Melvin V. Simpson at Tufts College Medical School, Boston. "Prior to that, there wasn't any understanding of how proteins got broken down. In fact, it was uncertain as to whether proteins turned over in normal cells."
Simpson's discovery that the degradation of proteins requires energy was surprising at the time, since most researchers had believed that, to the extent that protein degradation did take place, it would be an energy-releasing process. In the early 1970s, Hershko and coworkers corroborated Simpson's finding by showing that cellular protein degradation requires the ATP-dependent enzyme tyrosine transaminase.
Rose says he and his coworkers "began to investigate the possibility of finding an in vitro cell-free system that would show a requirement for ATP. I tried to do that for a number of years without success, because I had chosen cells that were much too complicated. This is an important factor. What you really want to do is get the right system if you're going to make some progress." In 1977, cell biology professor Alfred L. Goldberg and coworkers at Harvard Medical School succeeded by producing a cell-free extract from reticulocytes (maturing red blood cells) in which energy-dependent protein degradation occurred.
Hershko's group, now including Ciechanover, soon began using such extracts to study the degradation process. Separating the cell-free reticulocyte system into fractions caused a loss of its protein-degrading ability, and subsequent recombining of fractions restored it, leading the researchers to conclude that at least two components--and perhaps three, four, or more--were required for cellular protein degradation. "This was an extremely important discovery," Ciechanover says, "as it was the beginning of a new paradigm. Now we know that there are more than 1,000 components of the ubiquitin system." A key component of one of the purified fractions turned out to be a small heat-stable peptide called APF-1
SUBSEQUENTLY, "Avram went to do a sabbatical at [Rose's] lab," Ciechanover says. Rose was "a superb enzymologist" who was into enzyme mechanisms, including the way some enzymes use ATP, and "Avram thought Ernie would help him decipher the role of ATP in protein degradation."
This first stint led to a series of sabbaticals and summer sessions that Ciechanover and Hershko spent with Rose at Fox Chase Cancer Center. Early on, the researchers found that APF-1 could attach to proteins, and in 1980 they hypothesized that APF-1 labeling was a signal that led to degradation of the labeled pr otein by a downstream protease. A group led by University of Utah biochemistry professor Martin Rechsteiner, and later another led by Goldberg, found this protease to be a large protein complex called the proteasome.
Meanwhile, Gideon Goldstein of New York University School of Medicine and coworkers reported having identified and sequenced a peptide "probably represented universally in living cells," and they called it "ubiquitous immunopoietic polypeptide," subsequently shortened to "ubiquitin." Three Fox Chase postdocs--Keith D. Wilkinson, Michael K. Urban, and Arthur L. Haas--reported in 1980 that APF-1 was identical to ubiquitin. "It was an unusual situation," Rose recalls. "Postdocs don't usually publish papers by themselves, but that's how it worked out."
To be conjugated to protein substrates, ubiquitin first needs to be activated. "We isolated the three enzymes [called E1, E2, and E3] that activate ubiquitin and conjugate it to proteins," Ciechanover says, "and Ernie was instrumental in deciphering the biochemistry of all the activations."
"I don't know why I was put into the whole [Nobel Prize-winning] business myself, but I guess I deserve a little credit," Rose says, particularly for his work on the mechanism of the E1, E2, and E3 system. "I also did some work on isopeptidases, enzymes that take ubiquitin off proteins and recover it for reutilization." However, Ciechanover calls the isolation and mechanistic elucidation of the E1, E2, and E3 system "the core discovery" of ubiquitin-mediated proteolysis because it revealed the molecular details of how the whole process worked.

Others who made important discoveries about the ubiquitin system include cell biology professors Alexander J. Varshavsky of California Institute of Technology and Daniel Finley of Harvard Medical School. Varshavsky, Ciechanover, and Finley found clear evidence that ubiquitination is required for many proteins to be degraded in living cells and that ubiquitin-mediated proteolysis is the major selective proteolytic system in the cell. Varshavsky's group discovered the major physiological functions of the ubiquitin system--in the cell cycle, DNA repair, transcriptional regulation, and stress responses--and identified the first protein degradation signals, features of proteins that cause them to become targets of ubiquitin conjugation. Varshavsky's lab was also responsible for a number of key discoveries about the E1, E2, and E3 system.
Research on the biological effects of ubiquitin-mediated proteolysis has continued. For example, several groups have contributed to a better understanding of the role of protein degradation in processes like the cell cycle and transcriptional regulation.
On the day of the Nobel announcement, a Nobel Foundation interviewer asked Ciechanover if he had any advice for young students of science. He replied that one should select a research problem that's original--like protein degradation was in the 1970s, when the predominant attention of most protein biochemists was focused elsewhere. Then, "just believe in yourself and do it," he said.






Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter