Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Gene Editing
The next generation of genome editing is making big changes to DNA
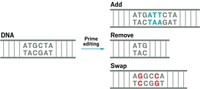
Using bridge RNA, scientists can induce large insertions, deletions, and rearrangements in a genome
by Max Barnhart
June 26, 2024
| A version of this story appeared in
Volume 102, Issue 20

The discovery of CRISPR-Cas9’s gene-editing prowess revolutionized genetic engineering just over a decade ago. Now it appears that genetic engineering technology may be taking its next big leap.
Two papers published in Nature on June 26 detail how bridge RNA adapted from a transposable element can induce large-scale, genomic changes at programmable and site-specific locations within a genome. Whereas CRISPR is best at making small, targeted modifications to a genome, bridge RNA gives genetic engineers a power they’ve never had before—to add, remove, invert, or rearrange large segments of DNA almost anywhere they want (DOI: 10.1038/s41586-024-07552-4 and 10.1038/s41586-024-07570-2).
Patrick Hsu, a bioengineer at the University of California, Berkeley, who led the research, calls bridge RNA the third generation of genetic engineering technology, after RNA interference and CRISPR. “It takes us beyond the DNA- and RNA-cutting abilities of CRISPR and RNA interference towards a broader suite of capabilities for the field of genome design,” he says.
That’s a strong claim, but one that other experts agree with. Yen-Ho Chen, a plant genome engineer working in industry, says, “It reminds me of when CRISPR was discovered. It’s novel, programmable, and you can tune this tool to adapt it for different applications. I think that part is potentially better than what we have right now in CRISPR-Cas9.”
The origin of the bridge RNA technique helps explain how it stands apart from RNA interference and CRISPR and is able to make these large genome modifications. Hsu and colleagues discovered bridge RNA when they were studying a transposable element in bacterial DNA called IS110. This genetic chunk can jump around the genome by cutting itself out of one spot and pasting itself in another.
IS110 produces a recombinase protein and expresses a noncoding RNA. The team found that the noncoding RNA contained two loops, a donor loop that could recognize IS110’s own DNA, and a target loop able to recognize DNA at an insertion site somewhere in the genome. When the RNA inside each loop bound to its respective DNA sequence, it formed a bridge, linking the DNA sequences together so that the recombinase protein could seamlessly merge them.
Hsu and the team realized they could reprogram the RNA in the donor and target loops to match any DNA sequence and therefore guide the IS110 recombinase protein to insert DNA, remove DNA, or make complex rearrangements at genomic locations of their choosing.
And that action of the recombinase proteinis what makes bridge RNA stand apart from CRISPR, according to Chen. CRISPR acts like scissors to cut DNA and then relies on the cell’s double strand break-repair system to fix the cut and insert a new gene. But this repair system is sometimes error prone and inefficient, meaning genetic engineers sometimes truggle to quickly get the result they want. Unlike CRISPR, the IS110 recombinase seamlessly combines the donor DNA with the target DNA and could therefore potentiallylet scientists insert DNA or make genomic rearrangements up to “millions of base pairs” long, Hsu says. That scale of genome modification is simply not feasible with CRISPR now.
But the technology for using bridge RNA in genome editing is still in the early stages. Hsu’s team has demonstrated this system only in bacteria, though Hsu says he is optimistic that efforts to adapt the approach to work in mammalian cells will succeed. Chen says this system’s efficiency may not be as good as CRISPR’s is now but that improvements will come with time.
Hsu says this technology “could one day modify entire sets of genetic variants simultaneously” instead of making gene modifications one at a time. He also believes bridge RNA might fix diseases caused by expansions of repetitive sequences in the genome like amyotrophic lateral sclerosis (ALS). “You can imagine repeat-expansion diseases like Huntington’s or ALS being collapsed by excising” the repeated sequences out of the genome,” Hsu says. Bridge RNA might also one day lead to cancer treatments by aiding in the development of cell and gene therapies, though such achievements are still some time away..



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter