Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Tracing Sars's Jump to Humans
Protein bound to its receptor reveals how virus adapted to human host
by Amanda T. Yarnell
September 19, 2005
| A version of this story appeared in
Volume 83, Issue 38

STRUCTURAL BIOLOGY
Public health experts hope that surveillance of animal coronaviruses for those poised to "jump" to humans could help prevent outbreaks of deadly diseases like severe acute respiratory syndrome (SARS). A new structural biology study has pinpointed what scientists should look for when patrolling for animal viruses that could cause another SARS outbreak.
The deadly human coronavirus behind the 2002-03 SARS epidemic--during which nearly 10% of the roughly 8,000 people infected died--originated in animals such as civet cats. Genetic sequencing has shown that the animal and human viruses differ by just a handful of mutations in a few viral genes.
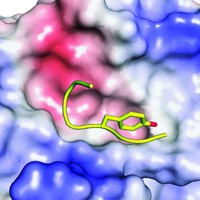
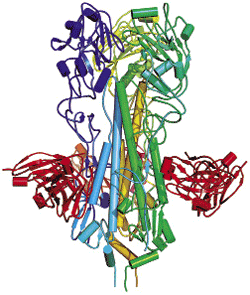
Some mutations are located on a spike-shaped protein that the coronavirus uses to bind to its host receptor, a zinc protease known as ACE2. A team led by structural biologist Stephen C. Harrison of Harvard Medical School has now revealed the structure of a piece of the human coronavirus spike protein bound to human ACE2 (Science 2005, 309, 1864).
"Understanding the structure of this complex will help us understand what these mutations in the spike protein mean in terms of infectivity," says Harrison, whose team includes his Harvard colleagues Fang Li, Wenhui Li, and Michael Farzan.
In the complex, the spike protein cradles ACE2. Smack in the middle of the interface between the two proteins is a pair of amino acids that Farzan's lab had previously fingered as being most critical for the virus's jump from animals to humans. These two amino acids differ from those found in the original animal spike protein, Harrison notes. The structure suggests that these mutations allowed animal-to-human transmission by increasing the affinity of the animal coronavirus spike protein for the human ACE2 receptor, he adds.
Harrison suggests that the structure might help scientists make predictive guesses about the potential infectivity of newly isolated viruses. He adds that the fragment of the spike protein used in the study is a promising starting point for the development of a vaccine against SARS.
His team also hopes to devise new SARS antivirals by studying the subsequent conformational changes in the spike protein that help usher the virus into its host cell. The Harvard team's search will join others now ongoing.
The spike protein is not the only promising target: A Chinese team recently reported the development of a class of peptidomimetic inhibitors of a protease conserved among many coronaviruses (PLoS Biol. 2005, 10, e324). They hope that the work will lead to a broad-spectrum drug that could be used to fight not only the SARS coronavirus but also those that cause conjunctivitis, bronchiolitis, and pneumonia.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter