Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Analyzing Protein Interactions
Technique probes inhibitor-enzyme interactions comprehensively
by Stu Borman
May 22, 2006
| A version of this story appeared in
Volume 84, Issue 21
Closely related enzymes often serve different cellular functions, but sorting out their distinct roles and the selectivity of their interactions with druglike compounds remains challenging. A new technique could accelerate basic research and drug discovery by probing sets of compound-protein interactions all at once, rather than in a tedious one-by-one manner.
Professor of cellular and molecular pharmacology Kevan M. Shokat and assistant professor of neurology William A. Weiss of the University of California, San Francisco, and coworkers devised the approach and used it on phosphoinositide 3-kinases (PI3-Ks) to elucidate insulin signaling and identify a promising brain-tumor inhibitor (Cell 2006, 125, 733 and Cancer Cell 2006, 9, 341).
The work "is remarkable in that it takes a fully fledged chemical genomics approach to the pharmacological targeting" of an enzyme family, comments Giulio Superti-Furga, director of the Center of Molecular Medicine at the Austrian Academy of Sciences, Vienna, on the Faculty of 1000 scientific review website. "The experimental cycle goes from the identification of new chemical entities that target the PI3-K family, to the structural basis for specificity among family members, to the use of selective compounds to assess biological function." The work "sets new standards in the use of chemistry to probe function of classes of targets, is a far cry from the majority of rather anecdotal findings on the subject, and exemplifies the power of an integrated interdisciplinary approach to compound selectivity."
Shokat, Weiss, and coworkers screened 55 kinases, including the entire PI3-K family, with a panel of 11 potent inhibitors and mapped the biochemical selectivities of each inhibitor for each target. They obtained crystal structures of representative inhibitor-enzyme combinations to understand the structure-activity relationships involved. They then carried out live cell and animal studies to determine the effects of specific interactions on insulin signaling and brain-tumor growth.
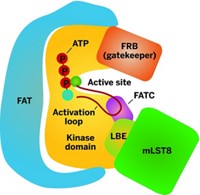
The insulin study revealed that PI3-K p110α is the primary control point for insulin signaling in cells, casting doubt on earlier findings. And the tumor study revealed that a compound called PI-103 blocks brain-tumor cell proliferation by interacting with two PI3-Ks: p110α and mTOR.
The new approach avoids the need "to piece together such insights from dozens of disparate papers that trickle out over years," says Shokat group postdoc Zachary A. Knight.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter