Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein Factory Reveals Its Secrets
Researchers picture and poke the ribosome to learn how it works
by Stu Borman
February 19, 2007
| A version of this story appeared in
Volume 85, Issue 8
Last year, the Nobel Prize in Chemistry heralded work on DNA transcription, a cornerstone process in molecular biology in which a cell synthesizes a messenger RNA (mRNA) version of genomic DNA. For some time, many research teams have been studying the other side of molecular biology's central dogma—the translation of mRNA into protein. That translation occurs on one of nature's most versatile molecular synthesizers: the ribosome.
If genomic DNA is the cell's planning authority, then the ribosome is its factory, churning out the proteins of life.
It's a huge complex of protein and RNA with a practical and life-affirming purpose-catalyzing protein synthesis. Bacterial cells typically contain tens of thousands of ribosomes, and eukaryotic cells can contain hundreds of thousands or even a few million of them. The ribosome found in the bacterium Escherichia coli is made up of three RNA components and more than 50 proteins. It weighs about 2.5 million daltons. Eukaryotic versions have four RNAs and about 80 proteins and weigh about 4 MDa.
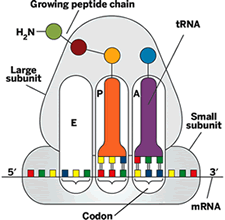
These dozens of components are all squeezed into two RNA-protein subunits, one small and one large. The ribosome's active site—where proteins are created by the one-at-a-time addition of amino acids to a growing peptide chain—is located in the large subunit. The active site may make protein, but it contains very little protein of its own, with only one of the ribosome's many protein components contributing to the mostly RNA architecture of the active site. Because RNA is so predominant in the active site, the ribosome is widely believed to be an RNA catalyst, or ribozyme—and, in fact, is thought to be the largest known ribozyme.
Understanding how the ribosome works is of fundamental interest, but such knowledge also could prove useful. For example, many antibiotics target bacterial ribosomes, so ribosome research could lead to new types of antibacterial agents. Researchers at the New Haven, Conn., start-up Rib-X Pharmaceuticals are riding on that hope. They have been using structure-based design in their efforts to discover novel ribosome-targeted antibiotics.
In the past decade or so, the ribosome has gone from being a biomolecule whose very structure was largely a mystery to one whose architecture is known at an atomic level and whose detailed workings are beginning to be better understood. Scientists have determined dozens of ribosome structures. They are conducting extensive mutational studies and are assessing the catalytic role of specific ribosome residues. They also have been carrying out theoretical modeling to aid understanding of the ribosome's detailed mechanism of action.
"The current model of [ribosomal] peptide bond formation is based on many different experiments, which sometimes did not seem to agree at first glance but little by little filled in the picture," says professor of physical biochemistry Marina Rodnina of Witten/Herdecke University, Witten, Germany.
Many basic facts about ribosome-catalyzed protein synthesis have long been known. The ribosome reads mRNA's genomic message and translates it into protein. When the ribosome factory is open for business, an mRNA binds to its small subunit, and amino acids corresponding to the mRNA's sequence are delivered one by one to the ribosome by aminoacyl transfer RNAs (tRNAs). Each tRNA molecule carries an anticodon, a three-nucleotide code that corresponds to the amino acid it's carrying. These anticodons must be matched up with corresponding codons (complementary three-nucleotide codes on mRNA), to enable a protein chain to be built to order.
The ribosome complex has three tRNA binding sites-A (aminoacyl), P (peptidyl), and E (exit). When peptide bond formation occurs, the amine from a new amino acid on the tRNA bound at the A site attacks a carbonyl at the end of the growing peptide chain, which is attached to the tRNA bound at the P site. The reaction lengthens the peptide by one amino acid unit.
As peptide bond formation occurs, the mRNA and tRNAs translocate-that is, they shimmy over one codon length. The P-site tRNA moves to the E site, where it gets ready to leave the ribosome, and the A-site tRNA moves to the P site, opening a space on the A site for a new tRNA. When the ribosome reaches an mRNA codon that signals a stop, the protein chain is released for use by the cell. The protein makes its getaway from the ribosome through a tunnel in the large subunit.
The ribosome doesn't carry out protein translation all by itself. It gets assistance from cofactors like EF-Tu, which delivers tRNA-amino acid complexes to the ribosome; EF-G, which catalyzes translocation; and release factors, which help synthesized proteins to exit the ribosome.
Over the past few decades, researchers have worked toward a much more detailed understanding of the way the ribosome works on an atomic level. Efforts go back at least to the 1960s, when Masayasu Nomura, now professor of biological chemistry at the University of California, Irvine, and coworkers showed that the ribosome could assemble spontaneously from its component RNAs and proteins. In the 1970s and '80s, Harry Noller, director of the Center for Molecular Biology of RNA at UC Santa Cruz, and coworkers used chemical modification studies to identify key ribosomal nucleotides and obtained evidence for the idea that translocation occurs in two discrete yet coupled steps.

Structural studies have spearheaded much of the progress since then in understanding the ribosome. Crystals of a ribosome subunit for crystallographic investigation were first made in 1980 by the late biochemist H. G. Wittmann of Max Planck Institute for Molecular Genetics, Berlin; structural biologist Ada E. Yonath of Weizmann Institute, Rehovot, Israel; and coworkers. Such crystals were initially prone to radiation damage. But in 1986, Yonath's group showed that the analysis of flash-frozen crystals, a technique called cryo-crystallography, can minimize radiation damage to the ribosome. This technique has improved the quality of data from subsequent crystallography of the ribosome and other biomolecules.
In the 1990s, Howard Hughes Medical Institute Investigator Joachim Frank of both the Wadsworth Center, Albany, N.Y., and the State University of New York, Albany, and his coworkers developed single-particle cryo-electron microscopy (cryo-EM) and began using it to study ribosome structure. Single-particle cryo-EM is a technique for imaging sets of individual molecules embedded in a thin layer of ice. Cryo-EM can be used to observe a greater variety of functionally interesting forms of biomolecules than is possible with crystallography. But cryo-EM can't normally attain atomic resolution, whereas X-ray crystallography can, so Frank and his coworkers often use crystallographic data to refine their cryo-EM maps.
By applying this combined approach to ribosomes, "we have been able to visualize a plethora of different processes that weren't seen before," Frank says. For instance, he and his coworkers showed how tRNA, upon entering the ribosome with EF-Tu, acts like a molecular spring by distorting as it contacts mRNA and then straightening as it enters the A site. They also found that when EF-G binds to the ribosome to induce translocation, the small and large subunits make a ratchet motion by rotating about 10 degrees about each other.
In 2000, several teams of researchers captured atomic (approximately 3-Å resolution) or near-atomic crystal structures of ribosome subunits. A Yale University group led by Thomas A. Steitz and Peter B. Moore reported a 2.4-Å structure of the large subunit, which continues to be the highest resolution ribosome structure of any kind. This study helped confirm that the ribosome is a ribozyme by showing that RNA predominates in the active site. The group subsequently obtained additional structures of the large subunit bound to various substrates, among them antibiotics and transition-state analogs.
Also in 2000, Yonath and coworkers obtained a structure of the small subunit and analyzed its structure with each of four antibiotics bound. The study revealed the antibiotics' binding sites and enabled the researchers to propose modes of action for these drugs. The team also obtained a structure of the large ribosomal subunit.
About the same time, structural biologist Venki Ramakrishnan of the MRC Laboratory of Molecular Biology at the University of Cambridge and coworkers obtained a crystal structure of the small subunit and also determined its structure with different antibiotics bound to it. Their study focused particularly on quality control of the decoding process, the way the ribosome checks codon-anticodon interactions between mRNA and newly arrived tRNAs.
The ribosome is strict about correct base pairing between the first two positions of three-nucleotide codons and anticodons. But it is more tolerant at the third; in fact, a handful of different synonymous codons (C&EN, Jan. 22, page 38) that differ only in their third positions can encode a single amino acid. The study revealed the structural basis for this redundancy in the genetic code. "We showed how the ribosome can discriminate between correct and incorrect tRNAs," Ramakrishnan says.
In 2001, a 5.5-Å resolution map of a whole ribosome with mRNA bound and tRNAs in the A, P, and E sites was obtained by Noller; Jamie H. D. Cate, now associate professor of chemistry, biochemistry, and molecular biology at UC Berkeley; Marat Yusupov of the Structural Biology & Genomics Laboratory, Strasbourg, France; and coworkers. The work revealed more about the relative orientation of the two subunits and their interactions with tRNAs. More recently, Cate and coworkers independently published a map of the whole ribosome, and Yusupov and coworkers independently obtained the structure of the ribosome with mRNA bound.
Last year, three more structures of the whole ribosome appeared. Ramakrishnan and coworkers obtained a 2.8-Å structure of the ribosome with mRNA and tRNAs bound. The study revealed that a kink in mRNA between the A and P sites is probably essential for maintaining the correct mRNA reading frame during translation. Noller and coworkers mapped the whole ribosome at 3.7-Å resolution with an mRNA mimic and two tRNAs in place. And Cate and coworkers obtained a 3.5-Å structure of the ribosome with mRNA and a tRNA mimic attached.
Such structural studies have opened the floodgates for a range of biochemical and computational research on the mechanism of action of the ribosome. "Lots of tidbits about the ribosome were out there already, but the structural work is what's 'crystallized' it all," says Rachel Green, professor of molecular biology and genetics at Johns Hopkins School of Medicine. "It's led to all the biochemistry that our group and several others have done on the ribosome."
For example, the idea that a specific RNA nucleotide in the ribosome active site accelerates peptidyl transfer by acting as a base was proposed by Steitz, Moore, molecular biophysics and biochemistry professor Scott A. Strobel, and coworkers at Yale. But results of mutational studies by Rodnina and coworkers and by Green's group contradicted that idea.
So did a subsequent study in which the entropy and enthalpy of the ribosome reaction were assessed by Rodnina; Richard V. Wolfenden, professor of chemistry, biochemistry, and biophysics at the University of North Carolina, Chapel Hill; and coworkers. If the peptide transfer reaction were base-catalyzed, it would be expected to have a large enthalpic component. But Rodnina, Wolfenden, and coworkers found that the origin of the 107-fold rate enhancement produced by the ribosome is entirely entropic and due to juxtaposition or desolvation of the substrates, not to base catalysis.
"The view accepted by most people now is that the active-site nucleotide does not play a dramatic role in peptidyl transfer," Green says. The general consensus, she says, is that orientation and positioning of substrates by the ribosome structure accelerates ribosome catalysis much more than any specific chemical effect.
Although the ribosome per se may not promote peptidyl transfer in a very active chemical manner, it's possible that the P-site tRNA substrate does catalyze the reaction-a proposed case of substrate-assisted catalysis. The growing protein chain is attached by an ester to a 3′-hydroxyl on one of the P-site tRNA nucleotide residues. In 2004, Strobel, Green, and coworkers confirmed earlier hints that peptidyl transfer is accelerated by a neighboring 2′-hydroxyl group on the same nucleotide. They found that deleting that 2′-hydroxyl causes a millionfold reduction in ribosome catalytic activity.
"The current model that everybody's discussing and likes is that that 2′-hydroxyl is essentially acting as a proton shuttle," Green says. The proton released from the A-site tRNA's nucleophilic amine apparently gets passed along to the 2′-hydroxyl. From there, it's passed to the protein's ester leaving group, which needs a proton to balance its negative charge. Strobel's group is currently carrying out experiments to further test the proposal.
Advertisement
A similar proton-shuttle mechanism had been proposed earlier by professor of theoretical chemistry Johan Åqvist of Uppsala University, in Sweden, and coworkers. They based their proposal on molecular dynamics and combined quantum mechanical/molecular mechanics simulations. Similar simulations by chemistry professor Arieh Warshel and coworkers at the University of Southern California, Los Angeles, support the proton-shuttle mechanism as well, although the USC group found that electrostatic stabilization-not substrate assistance or orientational entropy-accounts for most of the catalytic effect.
Computation and simulation are likely to be extraordinarily useful for further clarifying the ribosome's mechanism, because these techniques currently represent "the only way to study the ribosome in motion in atomic detail," says theoretical biologist Kevin Sanbonmatsu of Los Alamos National Laboratory. Sanbonmatsu and coworkers have used a supercomputer to simulate a working ribosome, identifying eight new potential antibiotic target sites. "The study is the largest simulation performed to date in biology," Sanbonmatsu says.

Last year, Yonath, chemistry professor Lou Massa of Hunter College of the City University of New York, crystallographer Jerome Karle of the Naval Research Laboratory, Washington, D.C., and coworkers turned to density functional theory to model ribosome catalysis. They reported a quantum mechanical transition state for peptide bond formation in the ribosome. The study also "defined the activation energy of the reaction and identified ribosomal interactions that seem to stabilize the transition state, which is formed while the A-site tRNA is rotating into the P site," Yonath says.
Such revelations notwithstanding, the ribosome continues to hold onto a few secrets. "There have always been researchers who think that we understand how the ribosome works," Noller says. However, "at this point, in spite of high-resolution crystal structures and decades of biochemical, genetic, and biophysical studies, I don't think we understand the fundamental mechanisms at all," he says,
"How do tRNAs and mRNA move during translocation, a process that involves molecular movements of many tens of angstroms every 50 milliseconds or so?" Noller asks. "What is the role of EF-G in that process? How does EF-Tu speed up binding of aminoacyl-tRNA to the A site by several thousandfold? The ribosome is enormous and tremendously conserved phylogenetically, yet the things we claim to understand at this point involve only a handful of nucleotides."
"The questions are getting finer, and they're also getting harder to ask," Strobel notes. "Where one person says we have the answers, the next person says we have the questions."




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter