Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
DNA's First Language
Behind every sequence of As, Ts, Cs, and Gs is a whole lot of molecular shape, and that's where DNA's business gets done
by Ivan Amato
May 18, 2009
| A version of this story appeared in
Volume 87, Issue 20

WHEN SECRETARY of Defense Donald Rumsfeld, in an unintentionally comedic justification of the U.S. invasion of Iraq during a press conference in May 2002, said "There are things we don't know we don't know," the media gave him a good tongue-lashing. But Elliott H. Margulies, a bioinformatics whiz with the National Institutes of Health's National Human Genome Research Institute (NHGRI), likes to summon that old newsbyte about deep ignorance as a reminder of how the genome remains a predominantly unexplored and unknown world. "It is so poignant in genomics right now," Margulies says in his office in Rockville, Md., just up the road from NIH's main campus.
It's a somewhat ironic point of view. After all, it was in 2003 that the scientific community for the first time had in hand essentially the entire nucleotide-by-nucleotide sequence of the human genome's 3 billion genetic letters—adenines (A), guanines (G), cytosines (C), and thymines (T). It was one of the most audacious raids on ignorance that humanity had ever accomplished.
But as is the way in science, this new knowledge begat new revelations of ignorance. For one thing, it revealed that the entire genome hosts only about 20,000 genes—about the same number as that of Caenorhabditis elegans, a dot-sized worm—rather than the 100,000-gene roster that scientists had so confidently bandied about for years with the presumption that more genes are better. It also showed that those protein-coding segments we call genes—long considered the genome's most biologically important content—occupy maybe 2% of the genomic landscape. That means 98% of that territory is doing something other than coding for proteins.
To begin exploring that vast molecular terra incognita, NIH launched in 2003 the Encyclopedia of DNA Elements Project (ENCODE), a large ongoing collaboration that includes Margulies, with the goal of uncovering all the components of the genome that have some biological function. In recent years, Margulies has had a nagging feeling that too much emphasis on the sequence of the genome's genetic letters could overshadow something even more fundamental and biologically relevant about genomes: the specific shapes that the DNA assumes at every genomic location.
In March, Margulies, chemist Thomas D. Tullius of Boston University, and colleagues there and at NIH's National Institute for Biotechnology Information unveiled in Science a tantalizing glimpse of what at least some of that uncharted genomic territory physically looks like (2009, 324, 389).
Rather than scanning only for evolutionarily conserved nucleotide sequences—usually represented by strings of letters, as in TCAATTTAGGAGCTTGC—that are common to many species' genomes and therefore likely to have biological functions such as coding for a gene, these scientists went hunting in genomes for common shapes or motifs embodied by short stretches of DNA. By adding this shape-driven way of scanning genomes to the traditional sequence-driven approach, the researchers reported seeing that twice as much genomic landscape is similar across mammalian species than a search for only similar strings of genetic letters reveals. In their Science paper, the researchers claim that 12% of the nucleotides in the human genome are evolutionarily conserved.
USING a shape-determining algorithm developed by Stephen Parker, a just-minted Ph.D. from Tullius' lab who will soon join Margulies in Rockville, the group analyzed the topographies of thousands upon thousands of 11-nucleotide stretches. That's a length that corresponds to a full twist of the double helix and happens to be convenient for the computational process of Parker's algorithm. The analyses revealed that DNA snippets with very different sequences can have very similar topographies and that snippets whose sequences differ by even a single nucleotide can assume dramatically different shapes. These morphological traits of DNA would not even get on the radar screens of researchers attuned only to sequences.
"Unfortunately, we bioinformatics guys think of DNA this way, as a string of letters," Margulies laments. He acknowledges that this letter-based representation of DNA has proven scientifically invaluable and exciting because it is effectively a digitized version of DNA that has rendered otherwise intractably large data sets—such as multi-billion-nucleotide genomes—amenable to rich and telling computer analyses, searches, and comparisons.
But this sequence-centrism has blinded researchers for decades to other ways of scanning DNA. After all, biological molecules do not see letters; they "see," "feel," respond to, and react with three-dimensional nucleotide motifs, rich with dynamic electrostatic, geometric, and other chemically determining traits. None of that is obvious in DNA when it is viewed as strings of letters.
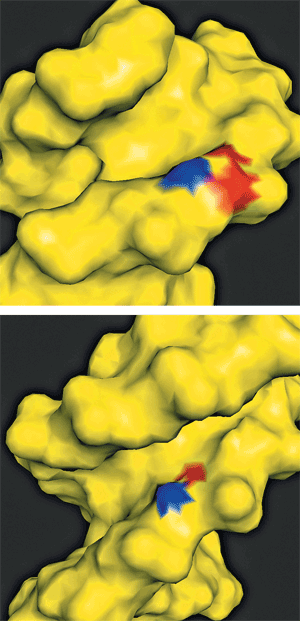
THIS IS WHERE Tullius' chemical skill set comes in. For many years, he has been developing and using a technique that relies on bond-busting hydroxyl radicals to shatter DNA into shards from which he can infer some of the overall shape of the intact DNA. The radicals form in solutions of iron(II)-EDTA (ethylenediaminetetraacetic acid) and hydrogen peroxide along with the DNA of interest. "The OH radical diffuses to the DNA and first runs into a sugar [on the DNA backbone in the minor groove], where it pulls off a hydrogen. HO∂ becomes water and leaves a radical at the position where it abstracted the hydrogen, and then those radicals react in a variety of ways that eventually give you strand breaks," Tullius explains.
Key to the technique is to fine-tune the cleavage reaction so that it shatters the DNA into stretches of about 100 nucleotides rather than all of the way down to its single nucleotides. By fluorescently tagging the fragments and then running them through an electrophoresis gel, the researchers can tell by the intensities of fluorescence of the different fragments just how the original piece of DNA shattered. And from that they can infer what they call a "structural profile of a DNA region." It's a technique, they say in their Science paper, "that can be used to predict the shape of the DNA backbone and grooves of genomic DNA at a single-nucleotide resolution."
"DNA sequence is not the be-all and end-all. Shape is primary."
The logic that connects the shatter pattern of DNA to its shape goes like this: The ability of the OH radicals to approach specific locations in a stretch of DNA corresponds to the "solvent accessibility" of that region, which corresponds to how wide or narrow DNA's minor groove is at the location. So from the glowing DNA shards that show up in the electrophoresis run, it is possible to reconstruct a portrait of the DNA that depicts the topographic variations in the minor groove (the smaller of the two grooves that wind along the DNA's helix; the other groove is known as the major groove) the way a map might show the variations in the width of a canyon.
The procedure shifts the focus from the genetic sequence to the structural motifs underlying those sequences. The challenge now is to connect those motifs and their specific chemically relevant shapes to the biologic roles they might be playing. Such potential functions include acting as selective binding sites for transcription factors and other proteins and nucleic acids involved in controlling genetic expression.
Tullius suspects he knows at least part of how these players find their specific binding locations in the vast genomic landscape. "Our patterns correlate really beautifully with the electrostatic potential of DNA's minor groove," he notes. Those nanoscapes of electric charge, in turn, can function like minuscule stretches of molecularly selective tape that grab onto specific proteins amid a diverse crowd of macromolecules in the nucleus. Says Tullius, "We are just starting to glimpse what is going on here."
It turns out that other researchers have observed this very phenomenon. Barry Honig, a Howard Hughes Medical Institute investigator at Columbia University, and his colleagues looked into how specific and structurally similar members of the Hox protein family—which orchestrates the development of the head-to-toe and some other gross morphologies of organisms ranging from fruit flies to mice to people—bind to the specific locations in the organisms' genomes. It is from these genomic perches that these transcription factors then contribute to the activation or silencing of specific genes.
"Our ignorance of DNA's information content is high," Honig says, reiterating that thinking and talking solely in terms of nucleotide sequences can't plumb the full richness that resides in our genomes.
BY WAY OF computational studies of structure, as well as in vitro and in vivo studies, Honig and his coworkers convinced themselves that the Hox proteins zero in on their proper locations by, in their words, "reading the structure and electrostatic potential of the minor groove" of DNA (Cell 2007, 131, 530).
It looks like a two-step process, Honig says. All Hox proteins, as they encounter DNA, are drawn to thymine- and adenine-rich regions by interacting with these bases in the DNA's major groove. But there are lots of these regions. To home in on particular ones, specific positively charged amino acids in the different Hox proteins insert into particularly narrow regions of DNA's minor groove, where the electrostatic potential is most attractive. These narrow regions are the same sort of motifs Margulies' and Tullius' group has been looking into at genome scales. The amino acids "that insert into the minor groove recognize a specific DNA structure" instead of the more general chemical cues provided by, say, the adenines and thymines, Honig and his collaborators explained in their Cell paper.
"Our ignorance of DNA's information content is high."
"The bottom line is that the width of the minor groove shapes the electrical potential, and this is being used in a very subtle way to discriminate very similar proteins," Honig tells C&EN. Without the ability for Hox proteins to land on and stick to specific DNA locations, the activity of the genome would not unfold in ways that orchestrate the proper development of basic bodily morphologies such as the number of segments in limbs or even the location of the head.
Nick Gilbert, a chromatin researcher at Edinburgh Cancer Research Centre, in Scotland, says he expects that a shape-centered approach to studying chromatin—the DNA and protein macromolecule that makes up chromosomes—is likely to uncover more of this genetic macromolecule's polymathic talents. As an example, he points to so-called supercoiling of chromatin by which these enormous linear molecules assume various strained topologies as they wind and contort into forms that fit inside of cells. These supercoiled shapes, Gilbert notes, store mechanical energy that he and others conjecture cells put to use to drive processes such as DNA replication or transcription to messenger RNA. "DNA sequence is not the be-all and end-all," Gilbert says. "Shape is primary."
As Tullius sees it, focusing on shape is getting back to DNA's chemical roots. "And if shape has the ability to provide information, which it does, nature will take advantage of it," he says. Were Rumsfeld to weigh in here about scientists' state of ignorance about DNA, he surely would say, "We know more about what we don't know."




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter