Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Human Genome Sequence Milestone
Health care improvements come into focus as human sequence marks its 10th year
by Stu Borman
June 21, 2010
| A version of this story appeared in
Volume 88, Issue 25
Happy birthday to the human genome sequence, or at least human knowledge of it. This week marks the 10th anniversary of the completion of the draft human genome sequence by the international Human Genome Project (HGP) and the private company Celera Genomics.
That artificial finishing point, announced on June 26, 2000, at the White House, has turned out to be a commencement. Since then, advances in collecting, organizing, and interpreting genetic data and dramatic reductions in the cost of gene sequencing have made it possible for genomics to have a growing influence on human health care.
It’s been said that a truly transformational technology will always have its immediate consequences overestimated and its long-term consequences underestimated, noted National Institutes of Health Director Francis S. Collins at a recent meeting. “I think that’s turning out to be true for what we are learning from the human genome. This science is driving a lot of the excitement right now in biomedical research, and that’s likely to continue for some time,” he said.
Collins, who led the U.S. component of HGP as director of the National Human Genome Research Institute (NHGRI) from 1993 to 2008, spoke at “The Human Genome: A Decade of Discovery, Creating a Healthy Future,” an anniversary symposium held in Washington, D.C., on June 7.
The cost of genome sequencing has plummeted so much over the past decade that companies are now marketing sequence data on disease risks directly to consumers. Collins recently tested this out by submitting his own DNA (under an assumed name) to three genotyping companies.
“All three of these companies seemed to have highly accurate lab methods in terms of their sequence data,” he said. “All three agreed that I had an increased risk of diabetes, maybe something like 50% higher than the average person. That was actually a bit of an eye-opener and caused me to change my health behaviors a bit. But they were all over the place in terms of other predictions, like prostate cancer, where one said [my risk] was lower than average, one said I was average, and the other said I was above average.
“That’s obviously unsettling if these are in fact recommendations that people are going to pay attention to,” Collins said. “There will be no doubt rather intense discussion over the course of the coming months over the degree to which this kind of direct-to-consumer marketing needs more oversight.”
In addition to the 10th anniversary of the draft sequence, it’s close to the 20th anniversary of HGP, which was launched in October 1990. There was no sequencing of the human genome in a serious way for about the first six years of the project. Pilot projects for human genome sequencing began in 1996, and 20 centers in six countries then worked together to generate the June 2000 draft sequence. Over the next three years, scientists continued to work on the draft sequence to improve its completeness and enhance its accuracy, and in April 2003, the high-quality reference human genome sequence was declared complete.
Improvements in sequencing technology eased the way to those sequences and are now beginning to affect human health care in ways “we could hardly have imagined 10 years ago,” Collins said. He noted that the cost of sequencing 1 million base pairs, which was about $20,000 in 1999, is now 20 cents and continues to drop. “There is no end in sight,” Collins said. “No laws of physics have to be violated for this to continue, and it’s continuing in a dramatic way.”
HGP’s human genome sequence cost about $400 million. But Illumina Inc. announced earlier this month that it is now charging $9,500 to sequence the genome of an individual with a serious medical condition. “The expectation is that we will reach the $1,000 genome, certainly in the next four to five years,” Collins said.
As genome sequencing has gotten cheaper, a variety of projects have been established to find out more about the genome and expand medical applications of genomics. For example, the Cancer Genome Atlas is currently being assembled “to understand at the most detailed molecular level for the 20 most common cancers what is driving them” and how that information can be used diagnostically and therapeutically, Collins said.
Encode, the Encyclopedia of DNA Elements, is collecting information on how proteins tell genes whether they should be on or off, on epigenetic (nonsequence) effects on gene function, and on how genetic variations contribute to disease.
The International Knockout Mouse Consortium is developing in stem cells a knockout of every single mouse protein-coding gene and then making those stem cells available for research. The aim is to improve the ability to determine the function of different working genes.
Researchers studying particular genes often want to obtain actual copies of those genes (called cDNAs), but it can take months of work to obtain each one. So the Mammalian Gene Collection is assembling and making available a complete set of cDNAs for both the human and the mouse.
The 1000 Genomes Project plans to sequence the genomes of at least 1,000 participants from different ethnic groups. It’s an international effort to establish a detailed catalog of human genetic variation to support future biomedical studies.
The Human Microbiome Project is creating a catalog of all the microorganisms living in and on our bodies to better understand how those microbes contribute to human health and disease.
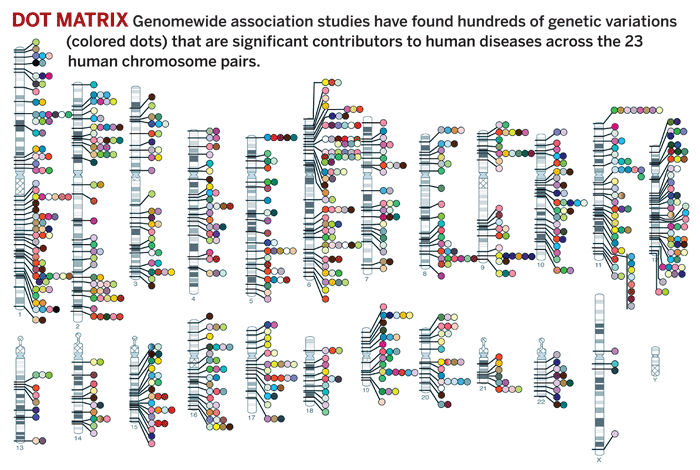
And the International HapMap Project is identifying the contributions of sequence variations to human disease. It focuses on the roughly 0.4% of the genome that differs among individuals—99.6% of different people’s gene sequences being identical—and on mapping these sequence variations to specific locations on chromosomes. HapMap data facilitate genomewide association studies, in which hundreds of genome-sequence associations with common diseases have now been validated. Many of these associations are weak or partial, but they nevertheless serve as pointers to potential drug targets.
Genomic data are also beginning to be used for personalized medicine—not only to provide the kind of data Collins obtained on his personal DNA but also to identify individualized responses to drugs. For example, in March, the Food & Drug Administration added a black-box warning to the anticlotting medication Plavix (clopidogrel) to warn patients and health care professionals that the drug is less effective in individuals with a variant gene for a liver enzyme that catalyzes formation of the drug’s active form.
Genomics will continue to get “closer and closer to patients” and will increasingly influence the way medicine is practiced, said the current NHGRI director, Eric D. Green, at the anniversary symposium. “You can just start to imagine all the projects that will spin out as you begin to sequence not dozens of human genome sequences, but hundreds, thousands, tens of thousands—because the technology is absolutely driving this.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter