Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Radical Start For Iron-Sulfur Enzyme
Novel protein’s 4Fe-4S cluster generates an unusual reactive species to make a modified amino acid
by Carmen Drahl
June 21, 2010
| A version of this story appeared in
Volume 88, Issue 25
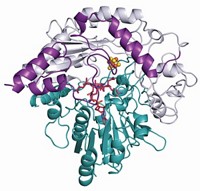
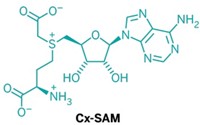
A modified amino acid that is the target of diphtheria toxin is assembled through some unusual enzyme reactivity, a multi-institution team has found (Nature 2010, 465, 891). Researchers have known for decades about the molecular target, called diphthamide. But its biosynthesis had remained unclear. Steven E. Ealick, Jack Freed, and Hening Lin of Cornell University in collaboration with Carsten Krebs of Pennsylvania State University and coworkers have revealed the structure and chemistry of a novel iron-sulfur enzyme that they say catalyzes the first step of diphthamide biosynthesis in a microorganism. Diphthamide, which is found on a protein factor used during translation, is a histidine residue modified with help from the cofactor (S)-adenosyl methionine (SAM). The team’s work suggests the new enzyme’s 4Fe-4S cluster transfers an electron to SAM to generate a 3-amino-3-carboxypropyl radical intermediate, which goes toward making diphthamide. Similar iron-sulfur enzymes all generate a 5´-deoxyadenosyl radical from SAM instead, so the new enzyme represents “a remarkable adaptation of hallmark reactivity,” Krebs says. Eventually, the team hopes to understand how many different enzymes generate different reactive species from SAM.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter