Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein Designers Roll Out A Barrel
Protein Design: After 28 years of failure, researchers have finally succeeded in creating from scratch a TIM-barrel, the most common protein fold found in enzymes
by Stu Borman
November 30, 2015
| A version of this story appeared in
Volume 93, Issue 47

The goal of de novo protein design is to understand protein architecture well enough to create new enzymes or other useful proteins from first principles, without modifying natural proteins. There have been many successes in the field, such as the design of globular proteins that fold with hydrophobic interiors and hydrophilic exteriors.
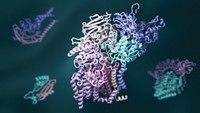
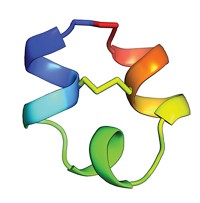
But the field has experienced at least one notable failure. As many as 10% of known enzymes adopt the cylindrically shaped triosephosphate isomerase-barrel (TIM-barrel) fold. For 28 years, protein designers have tried to create a TIM-barrel protein from scratch. But TIM-barrel design sequences have never folded into any tertiary structure—a three-dimensional assembly of α-helices, β-strands, and/or connecting loops—much less a TIM-barrel.
Now, a collaborative team reports that they have created the elusive barrel-shaped fold (Nat. Chem. Biol. 2015, DOI: 10.1038/nchembio.1966).
The researchers, led by Po-Ssu Huang and David Baker of the University of Washington, Seattle, and Birte Höcker of the Max Planck Institute for Developmental Biology, used rational design principles and calculations from the Baker group’s Rosetta protein modeling and design software to create sequences likely to fold into a TIM-barrel’s eight α-helices, eight β-strands, and connecting loops. They combined different features into an initial set of 22 sequences. They studied common elements in five sequences that folded well and zeroed in on a single prospect. The researchers then used X-ray crystallography to confirm that the sequence folded into a TIM-barrel conformation.
The work “represents a landmark achievement,” says macromolecule designer Deepesh Nagarajan of the Indian Institute of Science. “The implications are numerous,” including opening new avenues in enzyme design.
Indeed, the collaborative team now hopes to use designed barrel folds to create a new generation of custom-designed enzymes and as structural components of designed protein complexes, Huang says.





Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter