Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Process Chemistry
‘Magic mushroom’ enzyme mystery solved
Researchers unravel the biosynthesis of the psychoactive drug psilocybin, making large-scale production a possibility
by Stephen K. Ritter
August 14, 2017

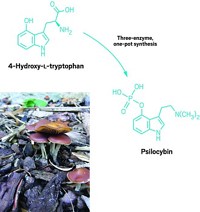
The euphoria and hallucinations induced from eating Psilocybe “magic mushrooms” have earned the fungi a cult following. Sandoz chemist Albert Hofmann isolated and determined the structure of psilocybin, the main ingredient in mushrooms that leads to the psychedelic effects, nearly 60 years ago. That discovery and subsequent mind-altering experiments by Harvard University psychologist Timothy F. Leary have left scientists longing to develop a large-scale synthesis of the compound for medical uses, which include treating anxiety and depression in people with terminal cancer and treating nicotine addiction. Yet no one has been able to unravel the enzymatic pathway the mushrooms use to make psilocybin, until now.
Janis Fricke, Felix Blei, and Dirk Hoffmeister of Friedrich Schiller University Jena have identified and characterized to the greatest extent so far the four enzymes that the mushrooms use to make psilocybin. The team then developed the first enzymatic synthesis of the compound, setting the stage for its possible commercial production (Angew. Chem. Int. Ed. 2017, DOI: 10.1002/anie.201705489).
During their study, Hoffmeister and coworkers sequenced the genomes of two mushroom species to identify the genes that govern fungal enzymatic production of psilocybin. They further used engineered bacteria and fungi to confirm the gene activity and exact order of synthetic steps. This process includes a newly discovered enzyme that decarboxylates tryptophan, an enzyme that adds a hydroxyl group, an enzyme that catalyzes phosphorylation, and an enzyme that mediates two sequential amine methylation steps. With that knowledge in hand, the team designed a one-pot reaction using three of the enzymes to prepare psilocybin from 4-hydroxy-L-tryptophan.
Medicinal chemist Courtney Aldrich of the University of Minnesota, Twin Cities, praises Hoffmeister and his coworkers for their painstaking efforts to elucidate the biosynthesis of psilocybin. “Our knowledge of the biosynthesis of fungal natural products has lagged behind our understanding of the corresponding bacterial biosynthetic pathways owing to a number of unique challenges,” Aldrich says. For instance, the genomes of fungi are more complex than bacteria, many fungi are still not amenable to genetic manipulation, and cultivating fungi to produce sufficient amounts of desired metabolites is not always straightforward. “The new work lays the foundation for developing a fermentation process for production of this powerful psychedelic fungal drug, which has a fascinating history and pharmacology,” Aldrich adds.
“The publication by Hoffmeister and colleagues highlights a terrific example of genomics-based biocatalyst-pathway discovery,” adds natural products researcher Jon S. Thorson of the University of Kentucky. “While psilocybin biosynthesis derives from a series of fairly simple chemical transformations, this new study identifies the contributing genes and biocatalysts for the first time and, importantly, provides strong evidence to support a revision of the order of the key steps proposed more than five decades ago. This work clearly sets the stage for bioengineered psilocybin production and/or for analogs that may serve as compelling alternatives to existing synthetic strategies.”
This article has been translated into Spanish by Divulgame.org and can be found here.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter