Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biochemistry
Newly discovered enzyme adds iodine to diverse substrates
Halogenase found in virus that infects photosynthetic bacteria
by Celia Henry Arnaud
October 15, 2019
| A version of this story appeared in
Volume 97, Issue 41

Halogenated compounds are widespread in the pharmaceutical and agrochemical industries. But the reagents used to make them can produce hazardous waste and mixtures of compounds. Enzymes can provide a milder, more specific route to halogenated compounds, but so far the options for adding iodine have been limited.
Now, a team led by Rebecca J. M. Goss of the University of St. Andrews has identified a viral halogenase they call VirX1, which adds iodine to diverse substrates with a preference for aryl species (Nat. Chem. 2019, DOI: 10.1038/s41557-019-0349-z).
“Although we once thought that halogens were rare in nature, over the past decade many halogenating enzymes have been reported,” says Sarah O’Connor, an expert on natural product biosynthesis at the Max Planck Institute for Chemical Ecology, who was not involved in the work. “However, iodinating enzymes have remained elusive.” Enzymes such as peroxidases can add iodine to substrates, but they have low selectivity.
Goss and her team were interested in new and diverse enzymes that mediate halogenation, particularly flavin-dependent halogenases, because they are known to be regioselective. They started their search for novel enzymes by looking for unique sequences that known halogenases share. They found a previously unnoticed motif in all known flavin-dependent halogenases and used it to search for new ones in diverse organisms.

Goss’s graduate student Danai S. Gkotsi, who found the viral halogenase, might have missed it. For the initial screen, Gkotsi used a simple chlorination assay because they expected the enzyme to be capable of chlorination or bromination. “Iodide salts have been shown to be inhibitors of flavin-dependent halogenases before, so direct screening with an iodide salt would have been too risky,” Goss says. But Gkotsi couldn’t detect any activity.
Goss and Gkotsi decided to try other halogen salts. “We were absolutely blown away when we saw that far and away the preference was for iodination,” Goss says. “And it wasn’t iodinating just one or two substrates. It was iodinating a really broad sweep of structurally very diverse compounds.” In a 400-compound library, they found 32 structurally and electronically very different compounds, including spirocycles, that the enzyme accepted as substrates. The enzyme, therefore, represents an exciting tool for late-stage diversification, Goss says.
VirX1 adds a single iodine to any given substrate. And it does it regioselectively, so there’s only one product rather than a mixture.
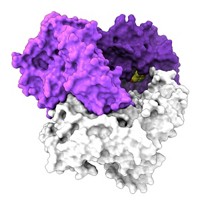
Crystal structures of VirX1 show that it’s a trimer with three identical subunits. The lack of a lid on its substrate-binding site, which is found in many other halogenases, may help explain its substrate diversity. The researchers were unable to get crystal structures of VirX1 with bound substrates, so they did molecular modeling studies instead.
The researchers found VirX1 in a virus that infects cyanobacteria. “It’s not just any old marine bacteria it’s going and infecting,” Goss says. “These are the most abundant photosynthesizers on the planet.” She doesn’t know VirX1’s role in the virus or what it does to the cyanobacteria host.
“This newly reported enzyme is a breakthrough in that it can preferentially and regioselectively install an iodo group on a variety of different substrates,” O’Connor says. “Given the importance of aryl iodides in many industrial processes, I can imagine that this enzyme is going to serve as a fantastic starting point for many new biocatalytic applications.”
Goss’s group is indeed working with industrial partners to see if they can selectively add iodine to pharmaceutical molecules. They are also trying to uncover the enzyme’s natural role.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter