Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Genomics
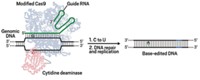
CRISPR-based nucleotide editor randomly changes RNA
Off-target editing remains a concern as scientists optimize CRISPR for human use
by Megha Satyanarayana
May 11, 2019
| A version of this story appeared in
Volume 97, Issue 19
Off-target effects have slowed the development of gene-editing technologies to treat human disease, and now researchers report that an editor they designed to make changes to DNA unexpectedly affects RNA as well. The scientific team, at Broad Institute of MIT and Harvard, reports that a base editor designed to modify specific adenosines out of DNA also randomly modifies adenosines in RNA (Sci. Adv. 2019, DOI: 10.1126/sciadv.aax5717). David Liu, the researcher who led the work, says they don’t know yet whether this indiscriminate activity has any biological effect, because RNA turnover in cells is very high. But some cancers can evade immune checkpoint inhibitors via errant adenosine-to-inosine editing similar to that seen in their study. Liu’s team sequenced RNA from cells trained to overexpress the adenosine base editor, called ABEmax, and compared the RNA with RNA from cells expressing mutant ABEmax and from cells without the editor. They found that about 0.22% of RNA was affected by normal ABEmax, versus 0.13% of RNA changed in cells without the editor. Cells make natural deaminases that convert adenosine to inosine. This difference is significant, Liu says, but cautions that it is still a small amount of off-target editing. The team is engineering ABEmax to be more specific to DNA.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter