Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Spectroscopy
Loren Andreas
Analysis ace is advancing methods for understanding proteins
by Celia Henry Arnaud
August 25, 2019
| A version of this story appeared in
Volume 97, Issue 33

Credit: Courtesy of Loren Andreas/Proc. Natl. Acad. Sci. U.S.A./ChemPhysChem/C&EN

Vitals
Current affiliation: Max Planck Institute for Biophysical Chemistry
Age: 35
PhD alma mater: Massachusetts Institute of Technology
Role model: “I credit Manish Mehta (Oberlin College) for sending me on my current scientific track, for lifting the curtain and exposing a world of spin physics behind NMR, and for showing it is possible to maintain scientific curiosity under the pressures of writing grants and publishing papers.”
If I were an element, I would be: “Proton/hydrogen. It has the strongest nuclear spin magnetic moment and is therefore the best detector nucleus for NMR.”
Must-have in the lab: “A solid-state NMR instrument with fast magic-angle spinning.”
Must-have on the road: “A backpack with a laptop and a toothbrush is enough. I try to leave the suitcase at home.”
3 key papers
“Structure of Fully Protonated Proteins by Proton-Detected Magic-Angle Spinning NMR” (Proc. Natl. Acad. Sci. U.S.A. 2016, DOI: 10.1073/pnas.1602248113)
“Structure and Mechanism of the Influenza A M218–60 Dimer of Dimers” (J. Am. Chem. Soc. 2015, DOI: 10.1021/jacs.5b04802)
“Dynamic Nuclear Polarization Study of Inhibitor Binding to the M218–60 Proton Transporter from Influenza A” (Biochemistry 2013, DOI: 10.1021/bi400150x)
Researchers have long wanted the ability to solve the structures of proteins in their natural state—in the complex environment of cells. But they’ve been stuck with methods that require them to study proteins in isolation. Loren Andreas hopes to change that using solid-state nuclear magnetic resonance spectroscopy to better understand biology and potentially open up new drug targets.
Andreas’s first big advance came as a graduate student in the labs of the Massachusetts Institute of Technology’s Robert G. Griffin. Using solid-state NMR, Andreas was able to solve the structure of part of influenza A’s M2 protein, a proton channel that goes through the virus’s envelope, or outer coat.
That work upended assumptions about the structure of M2. Previously, researchers thought the protein was a tetramer with fourfold symmetry, but it turned out to be a dimer of dimers. The findings have implications both for the mechanism of proton transport and for the binding site of inhibitors, which could be used to fight the flu. “That was his first home run,” Griffin says.
Andreas, working with Guido Pintacuda and Lyndon Emsley at the European Center for High Field NMR in Lyon, France, collected some of the NMR data for his PhD work using the center’s high-magnetic-field NMR instrument. In addition to working with 1 GHz magnetic fields, that lab is also pushing the frequencies used to spin samples in the magnetic field to speeds in excess of 100 kHz. That speed, twice as fast as is typically used in protein NMR, improves the resolution and sensitivity of these complex spectra.
After finishing his PhD, Andreas returned to Lyon for postdoctoral studies with Pintacuda. The combination of ultrahigh magnetic field and ultrafast spinning frequencies allowed them to solve structures of fully protonated proteins. Scientists typically use proteins in which many of the protons have been replaced with deuterium—a method that simplifies the NMR spectrum of the remaining protons. But not needing to deuterate the proteins makes sample preparation easier. The NMR community considers the 2016 paper reporting the fully protonated proteins a turning point in using NMR to study biological systems, Pintacuda says.
In his current work at the Max Planck institute for Biophysical Chemistry, Andreas continues to advance NMR of membrane proteins. He is studying a minimalistic β-barrel protein from oil-consuming bacteria that has long loops connecting the β strands. The role of those loops has been controversial: crystal structures of related proteins showed the loops as extensions of the β barrel, but solution NMR structures suggest more mobility in the loops. Andreas thinks his group will be able to resolve the controversy.
Andreas’s work “has opened what’s going to be an enormous field of research for NMR and for structural biology,” Griffin says. “He’s really opened a door for the rest of us to enter.”
Andreas doesn’t know what the next several years hold. By design, group leader positions at Max Planck institutes are not tenure track and are limited to a maximum of 9 years, so he’ll have to move on. He’d like to come back to the US to be closer to family. But he’s also excited about the advances in NMR that are happening in Europe. “The 1.2 GHz instruments should be installed within the next year,” he says. “This will be a huge leap for solid-state NMR especially.”
Researchers have long wanted the ability to solve the structures of proteins in their natural state—in the complex environment of cells. But they’ve been stuck with methods that require them to study proteins in isolation. Loren Andreas hopes to change that using solid-state nuclear magnetic resonance spectroscopy to better understand biology and potentially open up new drug targets.
Andreas’s first big advance came as a graduate student in the labs of the Massachusetts Institute of Technology’s Robert G. Griffin. Using solid-state NMR, Andreas was able to solve the structure of part of influenza A’s M2 protein, a proton channel that goes through the virus’s envelope, or outer coat.
Advertisement
That work upended assumptions about the structure of M2. Previously, researchers thought the protein was a tetramer with fourfold symmetry, but it turned out to be a dimer of dimers. The findings have implications both for the mechanism of proton transport and for the binding site of inhibitors, which could be used to fight the flu. “That was his first home run,” Griffin says.
Andreas, working with Guido Pintacuda and Lyndon Emsley at the European Center for High Field NMR in Lyon, France, collected some of the NMR data for his PhD work using the center’s high-magnetic-field NMR instrument. In addition to working with 1 GHz magnetic fields, that lab is also pushing the frequencies used to spin samples in the magnetic field to speeds in excess of 100 kHz. That speed, twice as fast as is typically used in protein NMR, improves the resolution and sensitivity of these complex spectra.
After finishing his PhD, Andreas returned to Lyon for postdoctoral studies with Pintacuda. The combination of ultrahigh magnetic field and ultrafast spinning frequencies allowed them to solve structures of fully protonated proteins. Scientists typically use proteins in which many of the protons have been replaced with deuterium—a method that simplifies the NMR spectrum of the remaining protons. But not needing to deuterate the proteins makes sample preparation easier. The NMR community considers the 2016 paper reporting the fully protonated proteins a turning point in using NMR to study biological systems, Pintacuda says.
In his current work at the Max Planck Institute for Biophysical Chemistry, Andreas continues to advance NMR of membrane proteins. He is studying a minimalistic β-barrel protein from oil-consuming bacteria that has long loops connecting the β strands. The role of those loops has been controversial: crystal structures of related proteins showed the loops as extensions of the β barrel, but solution NMR structures suggest more mobility in the loops. Andreas thinks his group will be able to resolve the controversy.
Andreas’s work “has opened what’s going to be an enormous field of research for NMR and for structural biology,” Griffin says. “He’s really opened a door for the rest of us to enter.”
Andreas doesn’t know what the next several years hold. By design, group leader positions at Max Planck institutes are not tenure track and are limited to a maximum of 9 years, so he’ll have to move on. He’d like to come back to the US to be closer to family. But he’s also excited about the advances in NMR that are happening in Europe. “The 1.2 GHz instruments should be installed within the next year,” he says. “This will be a huge leap for solid-state NMR especially.”
Vitals
Current affiliation: Max Planck Institute for Biophysical Chemistry
Age: 35
PhD alma mater: Massachusetts Institute of Technology
Role model: “I credit Manish Mehta (Oberlin College) for sending me on my current scientific track, for lifting the curtain and exposing a world of spin physics behind NMR, and for showing it is possible to maintain scientific curiosity under the pressures of writing grants and publishing papers.”
If I were an element, I would be: “Proton/hydrogen. It has the strongest nuclear spin magnetic moment and is therefore the best detector nucleus for NMR.”
Must-have in the lab: “A solid-state NMR instrument with fast magic-angle spinning.”
Must-have on the road: “A backpack with a laptop and a toothbrush is enough. I try to leave the suitcase at home.”
Research at a glance
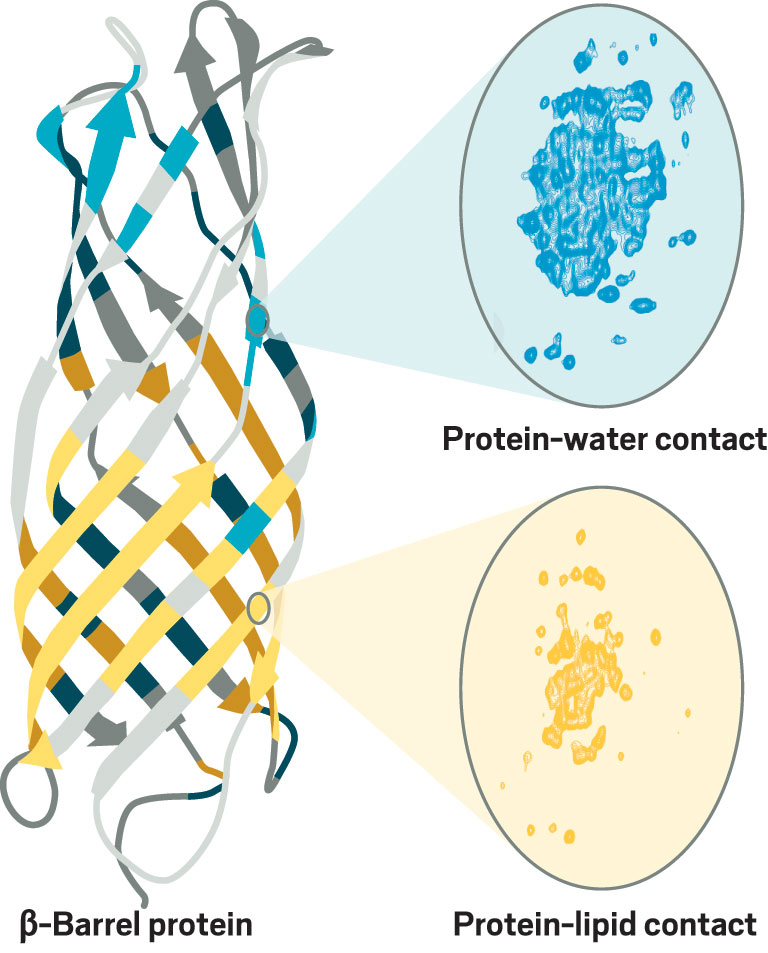
Credit: ChemPhysChem/Yang H. Ku/C&EN
Andreas combines high magnetic fields and ultrafast spinning frequencies to get better NMR spectra of membrane proteins. For example, he used NMR to obtain atomic-resolution maps of regions of a β-barrel protein in contact with water (blue) or lipids (yellow).
Three key papers
“Structure of Fully Protonated Proteins by Proton-Detected Magic-Angle Spinning NMR”
(Proc. Natl. Acad. Sci. U.S.A. 2016, DOI: 10.1073/pnas.1602248113)
“Structure and Mechanism of the Influenza A M218–60 Dimer of Dimers”
(J. Am. Chem. Soc. 2015, DOI: 10.1021/jacs.5b04802)


















Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter