Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Nature has been synthesizing organic molecules far longer than people have. Cells in plants, animals, microbes, and our own bodies perform countless chemical reactions, producing molecules called metabolites that can serve as organisms’ food, weapons, or waste. Metabolites are also useful to chemists. The molecules can serve as a starting point for new medicines, point out potential environmental toxins, and help diagnose cancers and cardiovascular disease.
Vitals
Current affiliation: Princeton University
Age: 29
PhD alma mater: University of British Columbia
Hometown: Victoria, British Columbia
If I were an element, I’d be: “Silicon because it plays central roles in both technology and biology.”
When I’m not doing chemistry, I enjoy: “Running, cycling, hiking, and backpacking. One of the best trips I’ve done recently was a backpacking trip in the remote Chilcotin mountains of British Columbia.”
But despite the myriad analytical tools at their disposal, scientists still struggle to understand metabolites and all that they do. We’re so unaware of “so much chemistry in the world around us,” Michael Skinnider says. At Princeton University, he’s combining the powers of mass spectrometry (MS) and machine learning to help shed new light on metabolites. It’s possible with mass spectrometry to detect every chemical in a complex sample, such as a tumor or a speck of soil, according to Skinnider. In other words, each molecule in the sample appears as a peak on the spectrum. But that’s not the same as knowing what each molecule is. “We can’t identify the metabolites,” he says.
Skinnider sees an opportunity to use machine learning to fill the gap between what we can detect and what we can name and draw a structure of. Machine learning tools, like large language models used in ChatGPT, can process huge amounts of information—such as MS data or molecular structures—and make accurate predictions about unknown molecules.
His plans go beyond identifying known and unknown metabolites. Skinnider hopes that as he and the other researchers in his group learn more about metabolites found in our bodies, they can start to link these molecules to cancers and other diseases.
Though Skinnider has ambitious scientific goals today, he didn’t even consider a career in science until the summer after his second undergraduate year at McMaster University.
He had spent the previous summer cleaning hotel rooms and making pizzas, and he wanted seasonal work that would be less physically taxing. So he took a gig with McMaster biochemist Nathan Magarvey coding computational tools to identify potentially interesting metabolites in large databases. Skinnider’s prior coding experience had been building websites for businesses in high school.
Within a few months, Skinnider says, “I was totally in love with it and wanted to do it the rest of my life.” While he’d enjoyed chemistry and biology classes, he found this work compelling because he was thinking about problems in ways no one had before.
By the time he graduated from McMaster 2 years later, Skinnider had cofounded a company with Magarvey that makes software tools to find new metabolites with potential applications in food, agriculture, and medicine. That firm, Adapsyn Bioscience, has received funding from Pfizer to search for potential drug candidates encoded in bacterial genomes and from the Bill and Melinda Gates Foundation to look for new tuberculosis treatments.
Advertisement
Always staying on top of things, Skinnider tracked down biochemist Leonard Foster to discuss research projects before entering an MD-PhD program at the University of British Columbia (UBC). Skinnider’s thesis work involved mapping protein-protein interactions in mouse tissue.
But that work wasn’t all that he accomplished at UBC. “I’ve never been very good at focusing on one thing,” he admits.
Foster gave him free rein to explore other research. One project led to a machine learning tool that could predict likely structures of new designer drugs even before law enforcement agencies were aware of them. The program learns known molecular structures of illicit drugs and then draws new structures that are likely to have similar properties and would be easy to synthesize. Drug enforcement agencies and toxicology labs in the US and Canada have reportedly implemented Skinnider’s tools.
That work, maybe more than any other, demonstrates Skinnider’s imagination, Foster says. “He’s thinking, ‘What can we do that is way beyond what anyone is thinking is possible at this time?’ ”
Skinnider started his own research group at Princeton barely a year ago. He admits it has been difficult—especially because he needs to stay a bit more focused than he’s used to. If he’s successful in marshaling the combined powers of mass spectrometry and machine learning, his group could soon shine light into some of the dark corners of the universe of metabolites.
Computational Chemistry
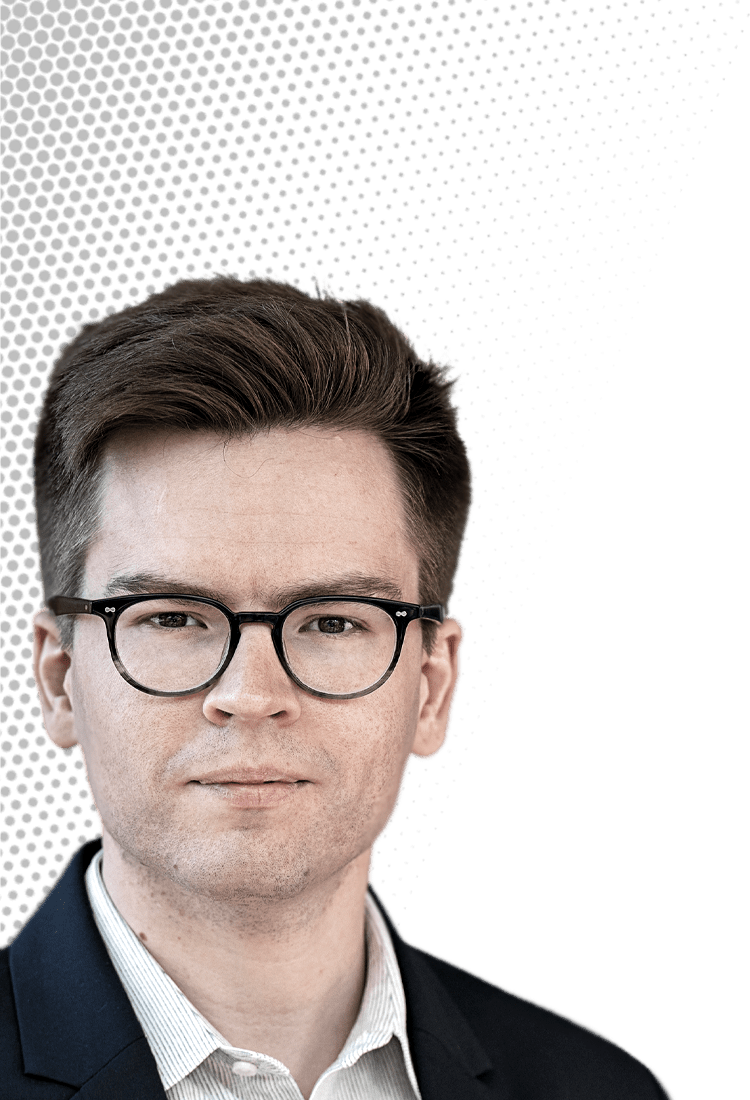
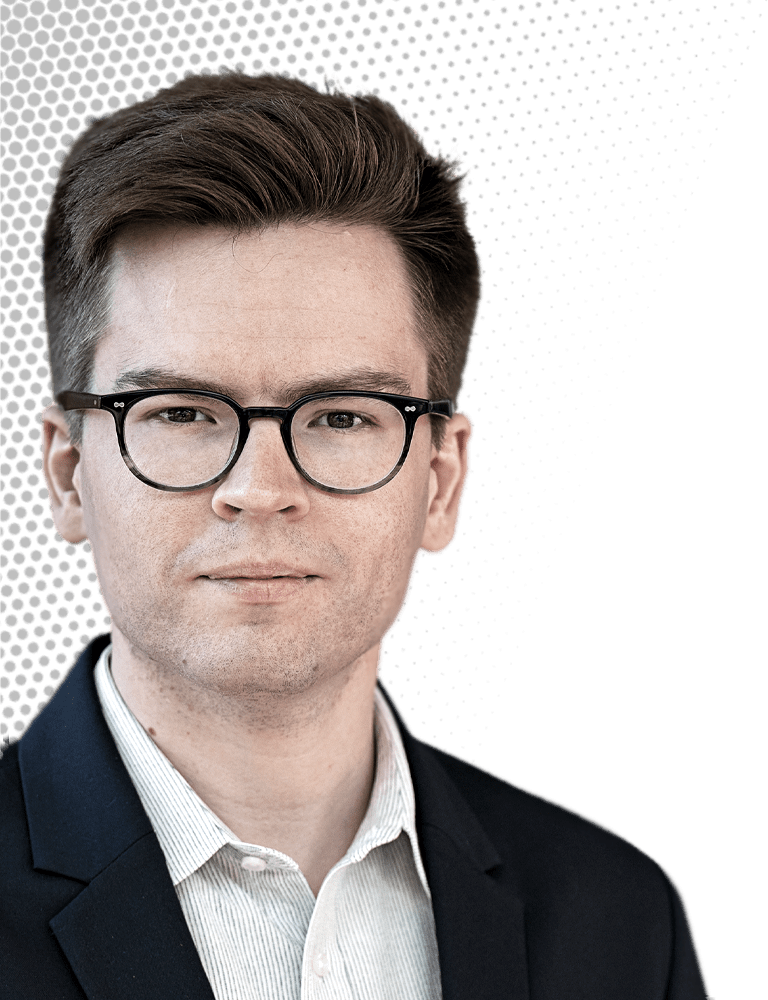
Michael Skinnider
This computational chemist combines mass spectrometry and machine learning to uncover metabolites
by Sam Lemonick, special to C&EN
May 17, 2024
| A version of this story appeared in
Volume 102, Issue 15

Credit: Sameer Khan/C&EN | Michael Skinnider
Vitals
Current affiliation: Princeton University
Age: 29
MD-PhD alma mater: University of British Columbia
Hometown: Victoria, British Columbia
If I were an element, I’d be: “Silicon because it plays central roles in both technology and biology.”
When I’m not doing chemistry, I enjoy: “Running, cycling, hiking, and backpacking. One of the best trips I’ve done recently was a backpacking trip in the remote Chilcotin mountains of British Columbia.”
Nature has been synthesizing organic molecules far longer than people have. Cells in plants, animals, microbes, and our own bodies perform countless chemical reactions, producing molecules called metabolites that can serve as organisms’ food, weapons, or waste. Metabolites are also useful to chemists. The molecules can serve as a starting point for new medicines, point out potential environmental toxins, and help diagnose cancers and cardiovascular disease.
But despite the myriad analytical tools at their disposal, scientists still struggle to understand metabolites and all that they do. We’re so unaware of “so much chemistry in the world around us,” Michael Skinnider says. At Princeton University, he’s combining the powers of mass spectrometry (MS) and machine learning to help shed new light on metabolites.It’s possible with mass spectrometry to detect every chemical in a complex sample, such as a tumor or a speck of soil, according to Skinnider. In other words, each molecule in the sample appears as a peak on the spectrum. But that’s not the same as knowing what each molecule is. “We can’t identify the metabolites,” he says.
Skinnider sees an opportunity to use machine learning to fill the gap between what we can detect and what we can name and draw a structure of. Machine learning tools, like large language models used in ChatGPT, can process huge amounts of information—such as MS data or molecular structures—and make accurate predictions about unknown molecules.
His plans go beyond identifying known and unknown metabolites. Skinnider hopes that as he and the other researchers in his group learn more about metabolites found in our bodies, they can start to link these molecules to cancers and other diseases.
Though Skinnider has ambitious scientific goals today, he didn’t even consider a career in science until the summer after his second undergraduate year at McMaster University.
He had spent the previous summer cleaning hotel rooms and making pizzas, and he wanted seasonal work that would be less physically taxing. So he took a gig with McMaster biochemist Nathan Magarvey coding computational tools to identify potentially interesting metabolites in large databases. Skinnider’s prior coding experience had been building websites for businesses in high school.
Within a few months, Skinnider says, “I was totally in love with it and wanted to do it the rest of my life.” While he’d enjoyed chemistry and biology classes, he found this work compelling because he was thinking about problems in ways no one had before.
By the time he graduated from McMaster 2 years later, Skinnider had cofounded a company with Magarvey that makes software tools to find new metabolites with potential applications in food, agriculture, and medicine. That firm, Adapsyn Bioscience, has received funding from Pfizer to search for potential drug candidates encoded in bacterial genomes and from the Bill and Melinda Gates Foundation to look for new tuberculosis treatments.
Always staying on top of things, Skinnider tracked down biochemist Leonard Foster to discuss research projects before entering an MD-PhD program at the University of British Columbia (UBC). Skinnider’s thesis work involved mapping protein-protein interactions in mouse tissue.
But that work wasn’t all that he accomplished at UBC. “I’ve never been very good at focusing on one thing,” he admits.
Foster gave him free rein to explore other research. One project led to a machine learning tool that could predict likely structures of new designer drugs even before law enforcement agencies were aware of them. The program learns known molecular structures of illicit drugs and then draws new structures that are likely to have similar properties and would be easy to synthesize. Drug enforcement agencies and toxicology labs in the US and Canada have reportedly implemented Skinnider’s tools.
That work, maybe more than any other, demonstrates Skinnider’s imagination, Foster says. “He’s thinking, ‘What can we do that is way beyond what anyone is thinking is possible at this time?’ ”
Skinnider started his own research group at Princeton barely a year ago. He admits it has been difficult—especially because he needs to stay a bit more focused than he’s used to. If he’s successful in marshaling the combined powers of mass spectrometry and machine learning, his group could soon shine light into some of the dark corners of the universe of metabolites.






Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter