Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Snapshots Of A Membrane Protein
First glimpses of cell-wall-forming enzyme will aid search for new antibiotics
by Stu Borman
March 12, 2007
| A version of this story appeared in
Volume 85, Issue 11
Antibiotic designers will get a boost from the first structural snapshots of an enzyme involved in forming bacterial cell walls. The coveted structures will assist rational design of molecules that can hit this promising but as-yet-untapped target.
Drug designers have long considered penicillin-binding proteins (PBPs) to be important drug targets. PBPs are dual-function enzymes that catalyze formation of a key bacterial cell wall component. One of the enzymes' domains, a transpeptidase (TP), is the target of β-lactam antibiotics such as penicillin. From structural studies of other TP enzymes, that interaction is understood. But many bacteria have developed resistance to this interaction, including methicillin-resistant Staphylococcus aureus, which causes infections in hospitals.
What has remained obscure is the way drugs interact with the other component of PBPs, the glycosyltransferase (GT) domain, which researchers would also like to use as a drug target. But structures of GTs of this type have not been available.
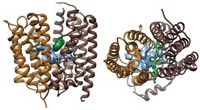
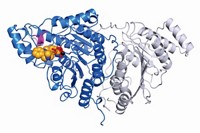
Now, two groups have addressed that problem. Professor of biochemistry and molecular biology Natalie C. J. Strynadka and coworkers at the University of British Columbia have obtained the first structures of a complete PBP, with and without an inhibitor bound to it (Science 2007, 315, 1402). Professor of microbiology and molecular genetics Suzanne Walker of Harvard Medical School and coworkers have obtained a higher resolution structure, but only of PBP's GT domain and without an inhibitor (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.0701160104).
The work could lead to new types of antibiotics and better versions of existing ones. The structures provide insight into "key interactions required for enzyme inhibition and critical information for the design of new drugs against these enzymes," Strynadka says.
From a fundamental perspective, the detailed mechanisms of action of PBP GTs proposed by the two groups are different. Both structures, however, show that the GT component adopts a fold distinct from those of all other GT classes. This finding is not entirely unexpected, as PBP GTs use lipid-sugar substrates, whereas the dozens of GTs analyzed previously all use nucleotide-sugar substrates.
The structures fill a critical knowledge gap about lipid-sugar GTs, which are found not just in bacterial membranes but are in fact "diverse enzymes that play many critical roles in cells of prokaryotes and eukaryotes," Strynadka says.
The inhibitor in some of the British Columbia group's structures is the antibiotic moenomycin, which is added to animal feed. Moenomycin is about 1,000 times more powerful than vancomycin, an antibiotic of last resort, but it is poorly absorbed, largely because it has a long lipid tail. "What's really promising is that now that we know how it binds the enzyme, we believe we can tweak moenomycin" for improved action in humans, Strynadka says.
"These enzymes are tremendously important antibiotic targets, and the co-complex with moenomycin, the only known inhibitor of the GT domain, is a major advance," Walker says of the other group's study.
Professor of chemistry and biochemistry Shahriar Mobashery of the University of Notre Dame, a specialist in bacterial resistance, says, "The picture of how bacterial cell walls are assembled is becoming clearer through the important contributions" of both groups.
And chemistry professor Gideon Davies of the University of York, in England, who specializes in structural enzymology, adds: "The results are insightful on many levels. This work will now inform rational design of new antibiotics."




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter