Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
CRISPR can now edit RNA
A new system called REPAIR targets and alters RNA bases, offering a more temporary alternative to DNA editing
by Ryan Cross
October 26, 2017
| A version of this story appeared in
Volume 95, Issue 43

As CRISPR gene editing marches closer to the clinic, several researchers have been tinkering to unleash the DNA editor on another nucleic acid: RNA, the intermediary messenger between DNA and proteins.
A team led by Feng Zhang of Broad Institute of Harvard University & MIT now report the first CRISPR complex engineered to edit individual bases of RNA (Science 2017, DOI: 10.1126/science.aaq0180).
“This is quite exciting, and I am talking about a competitor here, so I shouldn’t be excited,” says Mitchell R. O’Connell, who studies CRISPR RNA-targeting at the University of Rochester Medical Center. RNA editors will be valuable instruments for controlling gene expression in the lab, O’Connell says. And as a potential therapy, RNA editing “could be a safer alternative to gene editing,” because changes to DNA are permanent, but edits to RNA are temporary since cells make new RNA all the time, he adds.
The new study comes less than a month after another publication from Zhang’s lab showing that a CRISPR system with an enzyme called Cas13a can target and cleave specific strands of RNA (Nature 2017, DOI: 10.1038/nature24049). The classic CRISPR system uses an enzyme called Cas9 to cut DNA. Both systems require a programmable RNA strand, called a guide RNA, that directs the Cas enzyme to a complementary target—RNA for Cas13 and DNA for Cas9.
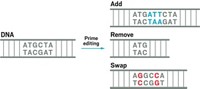
The new RNA editor effectively edits an adenosine (A) RNA base to guanosine (G). The method is based on yet another RNA-targeting enzyme called Cas13b. The team modified Cas13b so that it binds RNA but doesn’t cut it, and then they attached part of a second enzyme that converts an A nucleotide to the nonstandard nucleotide inosine (I). The cell’s protein synthesis machinery then reads the I as a G. Zhang calls the whole system RNA Editing for A to I Replacement (REPAIR).
The team showed that REPAIR could correct 35% and 23% of the mutations in the relevant RNAs for two diseases, X-linked nephrogenic diabetes insipidus and Fanconi anemia. O’Connell notes the success rate is likely to be even lower if used in an animal or human, but for some conditions, it may not be essential to fix 100% of the mutant RNA.
“It’s a tour de force,” says Dipali G. Sashital, who studies RNA-protein complexes, including CRISPR, at Iowa State University. “I think there is more work to be done, but essentially the idea that any RNA [sequence] could be targeted is pretty exciting.”
David B. T. Cox, a graduate student in Zhang’s lab, describes RNA editing as “a direct chemical conversion.” That’s an advantage over traditional DNA editing, Cox says, which requires cutting the DNA strand and could lead to undesired mutations. Another new CRISPR tool unveiled this week, called a DNA base editor, can change an A to a G also without snipping the DNA strand.
Zhang’s team already has a list of potential applications for REPAIR. For example, RNA editing could serve as a short-term therapy during wound healing and inflammation by modulating the activity or levels of RNA that produce proteins involved in those processes.
“Applications of the CRISPR system to RNA are heating up,” says Gene Yeo, a researcher at the University of California, San Diego, who developed a version of Cas9 to target and cleave RNA.
Yeo is a cofounder of Locana, a San Diego-based start-up developing therapies for conditions, such as amyotrophic lateral sclerosis, Huntington’s Disease, and myotonic dystrophy, involving long, repetitive stretches of RNA that result in the toxic buildup of proteins. Locana is using Cas proteins to target and snip these repetitive RNA stretches to hopefully treat the diseases. Earlier this year, Yeo demonstrated the concept in isolated human cells from a patient with myotonic dystrophy (Cell 2017, DOI: 10.1016/j.cell.2017.07.010).
“RNA targeting has many advantages, and I think this will grow much more because there are many more things you can do to RNA than DNA,” Yeo says.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter