Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biotechnology
CRISPR base editors cause unexpected mutations
A pair of studies show that cytidine base editors, but not adenine editors, create off-target edits
by Ryan Cross
February 28, 2019
| A version of this story appeared in
Volume 97, Issue 9

Papers from two independent groups suggest that a form of CRISPR gene editing, called base editing, causes a high number of unpredictable mutations in mouse embryos and rice.
It’s not the first red flag for CRISPR. Other groups have raised concerns about off-target mutations caused when the traditional CRISPR-Cas9 form of gene editing cuts DNA at a location that it wasn’t supposed to touch. The results of the new studies are surprising, however, because scientists have lauded base editors as one of the most precise forms of gene editing yet.
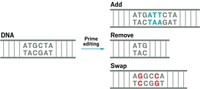
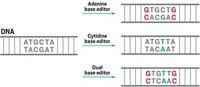
Base editors are designed to change one nucleotide, or letter, of DNA into another, without cutting the DNA. They come in two flavors: cytidine base editors, which convert a cytosine (C) nucleotide to a thymine (T), and adenine base editors, which convert an adenosine (A) to a guanosine (G). Both variants were created in David Liu’s lab at the Broad Institute of MIT & Harvard.
Dozens of labs have already published reports using the base editors in plants and animals, and Liu has cofounded two startup companies to commercialize the technology: Pairwise Plants and Beam Therapeutics.
Finding off-target mutations caused by CRISPR can be tough, because cells from different organisms, or even different cells from the same organism, can have slightly different DNA due to mutations that spontaneously arise over time. Unless the DNA between an edited cell and unedited cell were originally identical, it’s hard to know if any discrepancies were caused by CRISPR, or naturally-occurring mutations.
Hui Yang and a team of researchers from the Shanghai Institutes for Biological Sciences devised a clever technique to parse the two kinds of mutations. Using mouse embryos with only two cells—which should have identical DNA—they injected CRISPR-Cas9 or cytidine base editors into one cell, while leaving the other cell unedited. After the embryos grew for two weeks, they sequenced the DNA to compare the edited to unedited cells. The base edited cells had 20 times as many single-letter DNA mutations as the CRISPR-Cas9-edited and unedited cells. The results “surprised us all,” Yang says.

Yang recruited Lars Steinmetz, a geneticist at Stanford University, to help analyze the data. “I had anticipated that base editing might be a safer approach than CRISPR-Cas9 cutting,” he says. Then another twist emerged. Yang’s team repeated the experiment with adenine base editors, and found that they didn’t have the same problems (Science 2019, DOI: 10.1126/science.aav9973).
“I think these results raised great concern for the field of cytosine base editors,” Yang says. “Engineered base editors with higher specificities are needed to solve the problem.” His lab is already working on base editors that cause fewer off-target mutations.
Advertisement
A separate study, led by plant geneticist Caixia Gao at the Institute of Genetics and Developmental Biology in Beijing, found similar results when testing cytidine and adenine base editors in rice. Her team also discovered that the cytidine base editors still created mutations even when a crucial piece of the editor—the guide RNA—was missing (Science 2019, DOI: 10.1126/science.aaw7166).
CRISPR-Cas9 and base editors are both directed to their targeted site of editing by a guide RNA. A poorly chosen guide can create mutations at DNA sites with sequences that are closely, but not perfectly, complementary to the guide sequence. Scientists have improved all kinds of CRISPR editing by optimizing guide RNA design.
Steinmetz thinks there is a likely explanation for why cytidine base editors create mutations even when their guide RNA is withheld. Cytidine editors contain an enzyme called a cytidine deaminase, which converts a cytosine nucleotide into a uracil (U). During DNA replication or repair, that uracil, which doesn’t belong in DNA, is replaced with a thymine, resulting in the desired base edit. The cytidine deaminase could be acting on DNA alone, without following the direction of the base editor, Steinmetz suggests. It’s not a problem for the adenine base editor, which uses a different enzyme to make its edits.
“Base editing is in its infancy, so we are going to find stuff like this since it is so new,” says Alexis Komor, who designed the original cytidine base editor as a postdoc in Liu’s lab. Komor, who now runs her own lab at the University of California, San Diego, thinks this problem could be solved by engineering the cytidine deaminase to be bad at binding DNA on its own.
Another potential problem is the uracil DNA glycosylase inhibitor (UGI). Komor appended this inhibitor to the cytidine base editor to prevent cells from changing the uracil back into a cytosine. Uracil is actually a common mutation that spontaneously arises in DNA, so cells have a mechanism to quickly fix these mutations. Komor thinks it’s possible that the UGI on the cytidine base editor is blocking some of the cell’s normal repair processes.
The papers already have CRISPR scientists calling for a push to make better base editors. “The high off-target rates associated with cytidine base editors are surprising and alarming to us,” says Yinong Yang, a plant pathologist at Pennsylvania State University who has used CRISPR to design non-browning mushrooms. Yang says the new studies indicate that the specificity of cytidine base editors “needs to be significantly improved.”
Several groups have already published reports describing new versions of base editors, including Lukas Dow, a cancer biologist at Weill Cornell Medicine. Dow’s lab has improved the efficiency of the editor’s intended on-target editing, but not much work has been done yet to reduce the off-target editing, he says. He applauds the technical finesse of the new experiments, but adds that “there are still a lot of unknowns about how to interpret these papers going forward.”
For example, it’s not clear if the results of edits in mouse embryos will be similar to what happens when base editors are used in adult cells. Liu points out that the dose used in the new experiments was much higher than what would likely be used in therapies for humans. He also suggests that the mutation rate from the cytidine base editors may not be as high as they seem—around the same rate of mutations observed naturally in human cells. The risk of introducing these mutations will have to be weighed against the severity of the disease being treated, he says.
Beam Therapeutics, a startup that raised $87 million last year to create base editors for treating human diseases, thinks the issues presented in these paper are surmountable. “We are very confident that these are going to be medicines, and that we will be moving them forward to the clinic,” says John Evans, CEO of Beam. “That said, we have work to do still.”
Evans says that Beam already has 10 active drug discovery programs, using both cytidine and adenine base editors to treat genetic diseases. “We don’t view this finding as a unique roadblock to moving these base editors forward for patients, so we are very much still moving our pipeline forward,” Evans says.
The CRISPR field has had its share of scares, such as preclinical studies suggesting CRISPR could cause cancer or immune reactions. Some of these initial fears were overblown, while the importance of others won’t become clear until data from clinical studies rolls in. Gaétan Burgio, a scientist from the Australian National University who has watched CRISPR’s developments closely, thinks scientists could lose interest in base editors momentarily, but the problems may shake out over time. “The CRISPR field has this cycle of discoveries-obstacles-resolution,” he says. “Base editing is no exception.”
CORRECTION: This story was updated on March 27, 2019, to correct the name of the uracil DNA glycosylase inhibitor in the illustration of the cytidine base editor.
CORRECTION: This story was updated on March 1, 2019, to correct the spelling of Yinong Yang’s name.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter