Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Computational Chemistry
Covid-19
Computational scientists look for lessons learned from the COVID-19 pandemic
The pandemic changed these researchers’ methods and it showed them what works and what doesn’t during a global emergency
by Sam Lemonick
July 14, 2021
| A version of this story appeared in
Volume 99, Issue 28

The COVID-19 pandemic was an all-hands-on-deck event for scientists. Researchers of all stripes looked for ways they could help treat or stop the spread of the SARS-CoV-2 virus, and computational molecular scientists were no exception.
Support nonprofit science journalism
C&EN has made this story and all of its coverage of the coronavirus epidemic freely available during the outbreak to keep the public informed. To support us:
Donate Join Subscribe
Now, after nearly 18 months, the pandemic has entered a new phase in some parts of the world where vaccination rates are rising and the spread of the virus is slowing. Computational scientists are using the opportunity to take stock of their contributions to fighting the virus. They can point to times when the field’s efforts led to breakthroughs and also when those efforts fell short of expectations. But most importantly, they are trying to understand how their field can be more helpful when the next pandemic arrives.
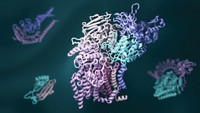
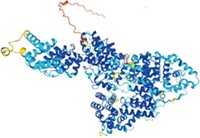
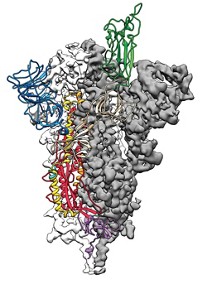
One computational achievement stands out from the rest—models of the virus’s proteins. In February 2020, with the virus spreading rapidly around the world, structural biologist Jason S. McLellan at the University of Texas at Austin and colleagues at the National Institutes of Health used cryo-electron microscopy (cryo-EM) to make detailed structures of SARS-CoV-2’s spike protein. The virus uses the spike protein to attach to and enter human cells. This protein is a major target for drugs and vaccines. Within weeks of McLellan’s team publishing the cryo-EM data, Rommie Amaro’s group at the University of California San Diego used those structures to create the first computer models of the protein using artificial intelligence and other computational techniques.
In the months that followed, the group used those tools to make more-highly-detailed models of the spike protein. For example, the researchers modeled what the sugars that dot the protein’s surface look like—a feature that cryo-EM can’t capture but that is important for understanding how antibodies or drugs may interact with the protein. Their simulations also showed how the protein’s shape changes to reveal its receptor-binding domain, a region that scientists want to target with therapeutics. The work won Amaro’s group a special kind of Gordon Bell Prize, one of the most prestigious awards in supercomputing. The Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research was created in 2020 to recognize pandemic-related research.

Amaro says the group was primed for success. In the same week that the cryo-EM structures of the spike protein were revealed, Amaro’s group published one of the most detailed computer models yet of the H1N1 influenza virus, one of the largest viruses to be modeled at that point. “We had the expertise ready to go” to target SARS-CoV-2, Amaro says.
Structural biologist Sarah Bowman of the Hauptman-Woodward Medical Research Institute says the models that Amaro’s group and others made at the beginning of the pandemic became part of an important back-and-forth effort between modelers and drug researchers: “People were depositing structures [in databases] so other people could say, ‘What therapeutics can we design?’ ” Amaro agrees that collaborations with experimental research groups have allowed groups like hers to refine their simulations with data from lab experiments, leading to more accurate models of the virus. She acknowledges that her models haven’t led to any drugs or vaccines yet, but she sees the work as the kind of basic science that may one day find use treating COVID-19 or another disease.
Another key to rapidly elucidating the virus’s structure and dynamics was access to supercomputers, Amaro and Bowman say. Amaro recalls seeing the first cryo-EM structures of the virus, which is even larger than the H1N1 virus. She called the executive director of the University of Texas at Austin’s supercomputing center and explained that SARS-CoV-2 was probably going to become a global problem and they’d need more computing resources to study it. “He said, ‘We’re going to let you have unfettered access,’ ” Amaro says. Her group ended up using about 250,000 of the Frontera supercomputer’s processors to make its initial model, more than twice the number of processors needed with its H1N1 model.
Other computational research efforts have not been as obviously productive as this structural work. Many scientists hoped that computational tools—whether established techniques like ligand-protein docking simulations or newer artificial intelligence methods—would quickly yield treatments for COVID-19. Those approaches have produced only one clear victory so far, baricitinib, and a small one at that.
In fact, the only drug that has been widely approved for treating COVID-19 was found without computational techniques. Researchers identified remdesivir, a broad-spectrum antiviral originally developed to treat hepatitis C, through lab experiments that evaluated the effects of several existing antiviral drugs on cells infected by SARS-CoV-2 (Cell Res. 2020, DOI: 10.1038/s41422-020-0282-0). This process of trying out already-approved drugs on new diseases is called drug repurposing.
Computers did play a central role in identifying another repurposing candidate, the arthritis medication baricitinib. Its utility was predicted in February 2020 by artificial intelligence algorithms developed and run by researchers at the company BenevolentAI (Lancet 2020, DOI: 10.1016/S0140-6736 (20)30304-4). The US Food and Drug Administration granted an emergency use authorization for baricitinib in combination with remdesivir in November. But neither remdesivir alone nor the two in combination have shown meaningful advantages for patients in terms of recovery time or survival rate (N. Engl. J. Med. 2020, DOI: 10.1056/NEJMoa2007764 and N. Engl. J. Med. 2021, DOI: 10.1056/NEJMoa2031994).

Computer-assisted drug design expert Alexander Tropsha of the University of North Carolina at Chapel Hill doesn’t see the lack of results as a failing of computational chemistry itself. Instead, he says that in some researchers’ rush to learn more about the virus or find potential cures, they did not take the time and care necessary for good results when they designed computational models and simulations. He notes that purely experimental efforts to repurpose drugs or find new ones have been similarly unsuccessful. In all cases, “What didn’t work was superficial,” he says. “What is beginning to work is high-level, rigorous, properly validated computational and experimental work in partnership.”
Recently, Tropsha and Artem Cherkasov of the University of British Columbia reviewed the efforts of computational scientists to look for COVID-19 treatments. They found that most were done without the necessary rigor in study design or expertise in interpreting results, producing hundreds of papers without rigorous results (Chem. Soc. Rev. 2021, DOI: 10.1039/d0cs01065k). Amid the early deluge of data on possible COVID-19 drug candidates, the preprint server bioRxiv stopped accepting computational studies of possible treatments unless they also included laboratory validation, apparently in an effort to keep low-quality or spurious results from reaching the wider media and the public. Pre-prints are scientific articles published before peer review as a way to share information more quickly.
Computational drug discovery expert Alpha Lee, a professor at the University of Cambridge and co-founder of the company PostEra, agrees with Tropsha about the quality of some early pandemic research. He says the easy, accessible computations some people did first are not very useful on their own. To make an impact during a pandemic, he says, experimental and computational scientists need to collaborate and share data. “Computational chemists can speed up the cycle and make things more efficient,” Lee says. “But without wet-lab work, saying, ‘I predicted 10 compounds active against COVID-19’ is not the best use of time.”
Lee’s company PostEra launched its own intense, open-science effort to find drugs to treat COVID-19, called the COVID Moonshot. Lee believes that this summer the scientists will have a molecule ready for lab tests that could lead to clinical trials.
While drug repurposing did find two treatments for COVID-19, no computational efforts have yet led to a new drug that can treat the virus. It’s not for lack of trying. In the first week of February 2020, the artificial intelligence company Insilico Medicine announced its algorithms had produced structures of new molecules that might treat the disease (ChemRxiv 2020, DOI: 10.26434/chemrxiv.11829102.v2). The company did another round of calculations before May and again in August. Despite the company’s prediction that drug trials involving patients might start as early as April 2020, none of Insilico’s molecules have yet made it into animal efficacy studies.
CEO Alex Zhavoronkov says the problem was a lack of resources, not the AI used in the molecule searches. He says the company couldn’t do its own laboratory evaluations of the molecules its AI predicted in the first several rounds, and the contract research organizations (CROs) they could have outsourced the work to were booked as the rest of the world raced to test possible COVID-19 drugs. In the second half of 2020, Insilico decided to build up its own laboratory capabilities, as it continued running its algorithms to find and refine potential drug structures. Zhavoronkov says that with its own resources and a growing availability of CROs as the worldwide pace of COVID-19 research slows, the company hopes to have one or two compounds in preclinical trials by the end of the year. That wait may even by a good thing: Zhavoronkov says each round of AI predictions has yielded more-promising molecules than the last.
In general, researchers say that these successes and failures of computational work during the pandemic have taught the field the value of a more open and collaborative way of doing science.
“I think what we learned from this pandemic is there is a need for sharing resources and information, and fast collaboration,” says computational chemist Joaquín Barroso-Flores of the National Autonomous University of Mexico. He and others point to a few ways computational research—and science more broadly—changed as the pandemic deepened, changes they say could help us tackle the next pandemic or address other threats, like climate change.
Some researchers made new agreements to publish data that they normally wouldn’t openly share. Amaro helped author one such pledge to share molecular simulations like hers, and structural biologists made their own pledge.
That willingness to share information as quickly as possible led to a rise in preprint postings. One study found that over 6,700 preprints related to COVID-19 posted between January and April 2020 and that scientists published results earlier and faster than before. On average, almost 400 times as many preprints came out each day during the COVID-19 pandemic as during the 2014 Ebola epidemic (Lancet 2021, DOI: 10.1016/S2542-5196(21)00011-5). The study’s authors point to differences between 2014 and 2020 that could have affected that rate—namely, the growth in the number of preprint servers. Traditional publishers are also increasingly willing to accept manuscripts that have been previously released as preprints.
Less-formal venues for sharing data and information also played an important part in COVID-19 research, Barroso-Flores says. “I was able to learn a lot from Twitter.”
He also highlights the increased access to supercomputing power like what Amaro used. Universities, government agencies, and corporations collaborated to make their computing resources more quickly and easily available to researchers working on COVID-19. The exact future of those partnerships is unclear, but some users have proposed making them permanent or finding ways to ensure the infrastructure that enables sharing computing resources stays intact for the next emergency. “That will be a legacy of this particular pandemic,” Amaro says.
Another thing researchers will need for the next pandemic is money, scientists say. “I hope governments will introduce new funding mechanisms” for emergency situations like COVID-19, Zhavoronkov says, so scientists will have the resources to quickly get to work.
Advertisement
The pandemic also forged valuable collaborations. Pharmaceutical companies including Novartis, Takeda Pharmaceutical, and Gilead Sciences saw the value in working together and creating an informal alliance to share resources and data that might help develop antiviral drugs. Takeda’s head of research, Steve Hitchcock, says his company has shared molecules and data about their properties with companies that are otherwise competitors. He thinks collaborating on precompetitive research—meaning work that won’t lead directly to a marketable drug—will stop companies from wasting time on redundant work.
And he thinks these efforts will continue beyond the COVID-19 pandemic. “I think there have been some permanent shifts in relationships. I think we realize we can do things faster together,” Hitchcock says.
For Barroso-Flores, cooperation was one of computational science’s biggest victories in the past 18 months. He knows that on its own, collaboration won’t be enough to tackle the next pandemic or other global challenges; scientists will need the kind of money and other resources that the COVID-19 pandemic unlocked. But he sees huge value in the human spirit of cooperation.
“At least we have proven that the human effort is there.”
Correction
On July 15, 2021, this story was updated to correct the description of the status of Insilico Medicine's testing of their molecules. The company has conducted some animal studies, but not animal efficacy studies.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter