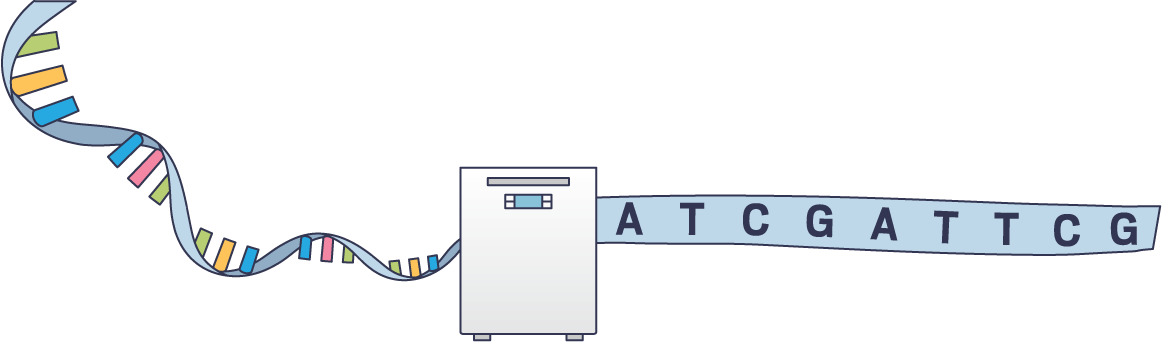
Sequencing
Credit: Shutterstock/Yang H. Ku/C&EN
Once researchers understood how DNA was shaped, they could begin to learn to read it. In 1977, biochemists reported a way to determine the sequence of a template strand. They used labeled bases to stop the replication of the template and determine which base was at each position (Proc. Natl. Acad. Sci. U.S.A., DOI: 10.1073/pnas.74.12.5463). That early method, known as Sanger sequencing, was used for decades for projects as varied as characterizing single genes and assembling the first maps of the human genome. Its successor, next-generation sequencing, hit the market in 2005. It has made DNA analysis faster and cheaper, enabling researchers to diagnose diseases rapidly, probe the differences in gene expression between individual cells, track pathogen evolution in real time, and catalog the diversity of microbial life. An ongoing challenge has been to enable more equitable access to the benefits of genomics. To yield benefits for all, research studies must include participants and scientists who represent not just populations with European ancestry but “the whole incredible tapestry of human genetic variation,” says Keolu Fox, a geneticist at the University of California San Diego.
Oligonucleotide synthesis

Credit: Shutterstock/Yang H. Ku/C&EN
In 1966, chemists reported a method to build an oligonucleotide with a desired sequence by anchoring the first base to a bead, then using protecting groups to add subsequent bases one at a time (J. Am. Chem. Soc., DOI: 10.1021/ja00974a053). The resulting short, defined oligonucleotides can be used as primers to start the polymerase chain reaction, which duplicates longer template sequences and became a key tool for molecular laboratories. Over time, researchers found efficient ways to produce long synthetic sequences by stitching together shorter ones; today it is possible to generate a whole synthetic genome this way (Nature 2023, DOI: 10.1038/s41586-023-06268-1). As drugs, synthetic oligonucleotides can suppress the expression of disease genes or introduce desirable proteins, such as in vaccines. Some engineers hope to use synthetic DNA to encode and store data as a greener alternative to servers.
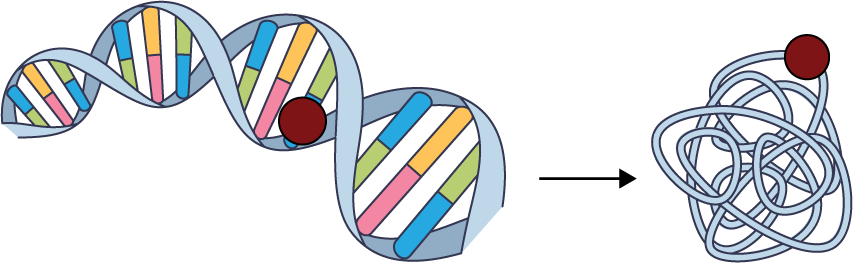
Protein engineering
Credit: Shutterstock/Yang H. Ku/C&EN
Once researchers understood how DNA encodes proteins, they could use it to change those proteins to suit their needs. Site-directed mutagenesis, a cloning method first reported in 1982, enabled researchers to pluck a single amino acid out of a protein and replace it (Nature, DOI: 10.1038/299756a0). Researchers used the technique to study proteins’ structures or tweak their activities. Today, engineered proteins are used as catalysts in industry, drugs in the clinic, and tools in the laboratory. Meanwhile, looking to a greener future, researchers use directed evolution to nudge proteins toward novel functions and artificial intelligence tools to design new proteins from scratch.
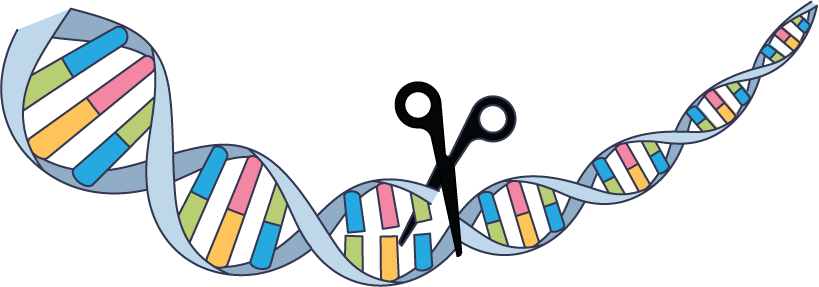
Genome editing
Credit: Shutterstock/Yang H. Ku/C&EN
Since researchers first harnessed bacterial immune proteins to manipulate sequences in living cells, CRISPR has spread rapidly into the public consciousness (Science 2012, DOI: 10.1126/science.1225829). Inspired by the protein Cas9, biologists have found and designed additional enzymes that can be guided to a target gene by complementary RNA. While delivery challenges remain for some medical applications, CRISPR-based genome editing has received US Food and Drug Administration approval to treat sickle cell disease (N. Engl. J. Med. 2021, DOI: 10.1056/NEJMoa2031054), is in tests for other genetic disorders, and has seen wide use in agriculture. The 2018 birth of twin girls whose genomes had been edited as embryos highlighted the urgency of ethical discussions about genome editing.
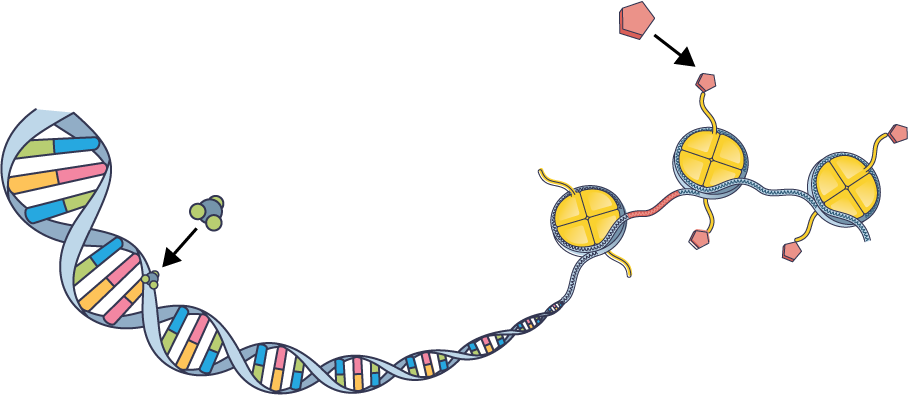
Epigenetic intervention
Credit: Shutterstock/Yang H. Ku/C&EN
Early in the 20th century, developmental biologists proposed multiple theories to explain how cells containing the same genes can commit to dramatically different fates. The discovery of DNA’s structure provided the foundation for exploring how changes unrelated to DNA sequence—either at specific points on DNA or across broad swaths of the genome—can modify gene expression. Today, researchers are becoming savvier about overlaying epigenetic and sequence data from single cells to decipher the biological basis of variation that’s not encoded in DNA. In the clinic, researchers are beginning to understand how to use epigenetic changes to predict disease progression or therapy response. Meanwhile, several cancer drugs target epigenetic enzymes.
DNA nanotechnology

Credit: Shutterstock/Yang H. Ku/C&EN
For nanoengineers, the double helix is just a starting point for creating more complex structures. With DNA as a building material, researchers can assemble tubes, lattices, and polyhedrons from short DNA tiles or fold longer strands into intricate 3D conformations. Researchers have built whimsical DNA structures, such as a Möbius strip or a tiny lockbox. They are also exploring practical applications, such as fabricating nanowires, etching microscopic nanoelectronics, and building tiny robots. Others hope to build DNA capsules for drug delivery and other therapeutic purposes, moving the field from engineering back toward the life sciences. “We steal material from biology, and we return the favor back to biology,” says Hao Yan, director of the Center for Molecular Design and Biomimetics at Arizona State University.














Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter