Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Computational Chemistry
Abigail Dommer
This molecular modeler wrangles billions of atoms to figure out how aerosols spread disease
by Laura Howes
July 15, 2022
| A version of this story appeared in
Volume 100, Issue 25

Credit: Will Ludwig/C&EN/Tim Peacock/Abigail Dommer
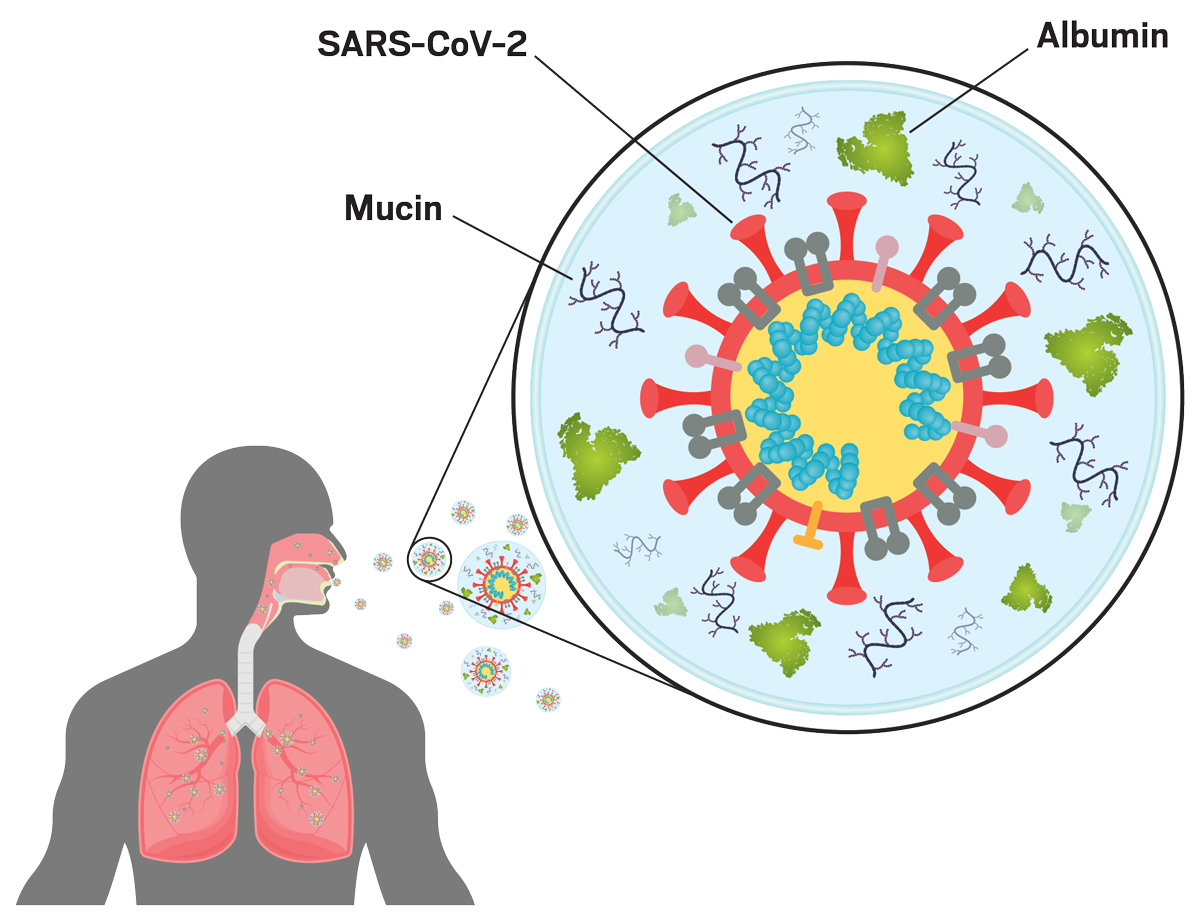
Maybe you saw the video on the New York Times website—a rippling sphere with ragged posts sticking out on thin stalks. It was an atomic-level model of SARS-CoV-2, the virus that causes COVID-19. Abigail Dommer was one of the University of California San Diego scientists who created that model. A PhD student at the time, she and her team modeled the coronavirus inside a respiratory aerosol, a supercomputing tour de force that calculated the interactions of over 1 billion individual atoms.
The SARS-CoV-2 model has been just one of the enormous molecular puzzles Dommer has solved computationally. These projects could help scientists understand how viruses move from person to person through the air or how sea spray induces clouds to form.
Advertisement
Dommer’s path to supercomputing superstar hasn’t been linear. She started her PhD in Kimberly Prather’s atmospheric chemistry and oceanography lab at UC San Diego before leaving graduate school to teach middle and high school science. She also learned to code and taught chemistry at local community colleges before deciding to return in 2017 to finish her PhD, after 2 years away.
That’s when Dommer, on the hunt for a new graduate supervisor, walked into the office of Rommie Amaro. Amaro’s lab builds complex computational models of biological systems like the flu virus. But Dommer was still interested in the ocean—in particular, sea spray.
Sea spray aerosols are microscopic particles ejected into the air by breaking waves. They influence our climate in different ways, and they are chemically complex—full of lipids, bacteria, viruses, and other gooey molecules that sit on the ocean’s surface. Dommer wanted to build big computational models of sea spray aerosols to understand how they affect climate.
Amaro said yes. Amaro couldn’t have predicted it when she accepted Dommer into her lab, but Dommer’s expertise in building ultralarge models of lipids in aerosols became invaluable when the COVID-19 pandemic hit in 2020. Amaro’s lab set itself the challenge of modeling the new coronavirus.
To model the whole virus, the team needed to slot the infamous spike proteins and other viral proteins into a spherical bilayer of floppy, jiggly lipids. Amaro naturally turned to Dommer for her expertise in modeling lipids in sea spray. Dommer was determined to make the simulation completely accurate. “I told Rommie, ‘I’m not doing this unless we can do it right,’ ” she says. That meant a year and a half researching the virus’s lipid composition and then developing a way to build a computational model without restraining the interactions of the molecules, which could limit the insights scientists could take from the model.
Solving the problem was frustrating, Dommer admits, but also satisfying. When she finally got a stable viral membrane, the team used it to build a 305-million-atom simulation of the whole virus. That model landed the team a slew of attention, including an ACM Gordon Bell Prize, sometimes referred to as the “Nobel for supercomputing.”
But Dommer was already thinking of the next puzzle—the aerosolized virus. What is the composition of those aerosols launched by people breathing and talking? And how does that composition affect transmission and infectivity? To answer those questions, Dommer built possibly the largest atomic simulation to date, modeling over 1 billion atoms.
“It’s so difficult to actually overstate her contributions,” Amaro says. “Abby managed to do multiple seriously amazing technical feats under intense time pressure, and then she created stunning images which captured the world’s imagination.”
Dommer completed her PhD work and is staying on at the Amaro lab as a postdoc to finish her work on aerosols. As she continues, she sees more links between respiratory mucus and marine gels inside the sea spray she studied.
“Every experience has given me something really new and has informed every other decision I’ve made,” she says. “I really like the feeling I’m building up a really colorful toolbox.”
Vitals
Current affiliation: University of California San Diego
Age: 30
PhD alma mater: University of California San Diego
Hometown: Columbus, Ohio
If I were an element, I'd be: “Bismuth. It creates the most beautiful geometric crystals and then upon surface oxidation becomes wildly iridescent. I’d be bismuth because adversity makes me more beautiful.”
My alternate-universe career: “I’ve always wanted to be the person who illustrates the infographics at zoos and botanical gardens.”
See the Talented 12 present their work at a virtual symposium Sept. 19, 20, and 21. Register for free at cenm.ag/t12symposium.
|
|
Maybe you saw the video on the New York Times website—a rippling sphere with ragged posts sticking out on thin stalks. It was an atomic-level model of SARS-CoV-2, the virus that causes COVID-19. Abigail Dommer was one of the University of California San Diego scientists who created that model. A PhD student at the time, she and her team modeled the coronavirus inside a respiratory aerosol, a supercomputing tour de force that calculated the interactions of over 1 billion individual atoms.
Vitals
▸ Current affiliation: University of California San Diego
▸ Age: 30
▸ PhD alma mater: University of California San Diego
▸ Hometown: Columbus, Ohio
▸ If I were an element, I’d be: “Bismuth. It creates the most beautiful geometric crystals and then upon surface oxidation becomes wildly iridescent. I’d be bismuth because adversity makes me more beautiful.”
▸ My alternate-universe career: “I’ve always wanted to be the person who illustrates the infographics at zoos and botanical gardens.”
The SARS-CoV-2 model has been just one of the enormous molecular puzzles Dommer has solved computationally. These projects could help scientists understand how viruses move from person to person through the air or how sea spray induces clouds to form.
Dommer’s path to supercomputing superstar hasn’t been linear. She started her PhD in Kimberly Prather’s atmospheric chemistry and oceanography lab at UC San Diego before leaving graduate school to teach middle and high school science. She also learned to code and taught chemistry at local community colleges before deciding to return in 2017 to finish her PhD, after 2 years away.
That’s when Dommer, on the hunt for a new graduate supervisor, walked into the office of Rommie Amaro. Amaro’s lab builds complex computational models of biological systems like the flu virus. But Dommer was still interested in the ocean—in particular, sea spray.
Sea spray aerosols are microscopic particles ejected into the air by breaking waves. They influence our climate in different ways, and they are chemically complex—full of lipids, bacteria, viruses, and other gooey molecules that sit on the ocean’s surface. Dommer wanted to build big computational models of sea spray aerosols to understand how they affect climate.
Amaro said yes. Amaro couldn’t have predicted it when she accepted Dommer into her lab, but Dommer’s expertise in building ultralarge models of lipids in aerosols became invaluable when the COVID-19 pandemic hit in 2020. Amaro’s lab set itself the challenge of modeling the new coronavirus.
To model the whole virus, the team needed to slot the infamous spike proteins and other viral proteins into a spherical bilayer of floppy, jiggly lipids. Amaro naturally turned to Dommer for her expertise in modeling lipids in sea spray. Dommer was determined to make the simulation completely accurate. “I told Rommie, ‘I’m not doing this unless we can do it right,’ ” she says. That meant a year and a half researching the virus’s lipid composition and then developing a way to build a computational model without restraining the interactions of the molecules, which could limit the insights scientists could take from the model.
Solving the problem was frustrating, Dommer admits, but also satisfying. When she finally got a stable viral membrane, the team used it to build a 305-million-atom simulation of the whole virus. That model landed the team a slew of attention, including an ACM Gordon Bell Prize, sometimes referred to as the “Nobel for supercomputing.”
But Dommer was already thinking of the next puzzle—the aerosolized virus. What is the composition of those aerosols launched by people breathing and talking? And how does that composition affect transmission and infectivity? To answer those questions, Dommer built possibly the largest atomic simulation to date, modeling over 1 billion atoms.
“It’s so difficult to actually overstate her contributions,” Amaro says. “Abby managed to do multiple seriously amazing technical feats under intense time pressure, and then she created stunning images which captured the world’s imagination.”
Dommer completed her PhD work and is staying on at the Amaro lab as a postdoc to finish her work on aerosols. As she continues, she sees more links between respiratory mucus and marine gels inside the sea spray she studied.
“Every experience has given me something really new and has informed every other decision I’ve made,” she says. “I really like the feeling I’m building up a really colorful toolbox.”




















Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter