Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Materials
First Amyloid Crystal Structures Were Just The Beginning
Since 2005, researchers have not only solved structures of many amyloid-forming peptides but also designed inhibitors
by Celia Henry Arnaud
December 21, 2015
| A version of this story appeared in
Volume 93, Issue 49
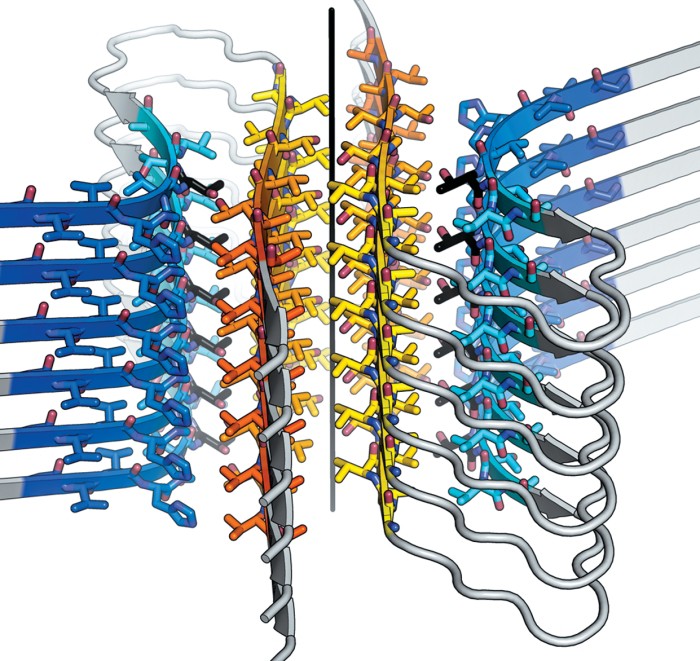
Scientists used to think it wasn’t possible to crystallize and therefore learn the structure of amyloid fibrils, the protein complexes such as amyloid-β and α-synuclein that are involved in Alzheimer’s and Parkinson’s diseases, respectively. But David S. Eisenberg of the University of California, Los Angeles, and coworkers proved such thinking was wrong. In 2005, they crystallized and solved the structure of a segment of amyloid fibrils—called the cross-β spine—that holds the complexes together (Nature 2005, DOI: 10.1038/nature03680).
“We learned that the adhesive segments of the proteins that form these fibers are just short segments,” Eisenberg says. The researchers didn’t need to get the whole fiber to crystallize as long as they had the adhesive portion. They could then work with synthetic peptides instead.
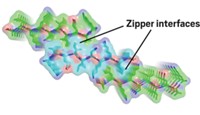
The crystals formed by the segments were too small for conventional X-ray crystallographic methods. But synchrotron methods had luckily advanced enough that Eisenberg and his colleagues could analyze the tiny crystals. The first crystal structures revealed that the fibril spine consists of two β-sheets with interdigitating amino-acid side chains that form a zipperlike structure.
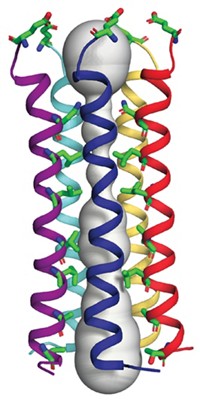
And those structures were just the beginning. Eisenberg’s team later showed that the steric zipper is a common structural motif in amyloid fibrils (Nature 2007, DOI: 10.1038/nature05695). In the past 10 years, they have solved the structures of the adhesive sections of many other disease-related proteins. This year, they solved the structure of the adhesive segment of α-synuclein, which had eluded them because it forms even smaller crystals than other segments do (Nature 2015, DOI: 10.1038/nature15368).
For Eisenberg, the crystal structures serve only as a jumping-off point. His team uses the atomic-level detail to design inhibitors that block fibril formation. Such inhibitors are typically peptides that bind to the top of a fibril and stop its growth. The first inhibitors were against tau, an Alzheimer’s-related protein, and SEVI (semen-derived enhancer of viral infection), a peptide that accelerates HIV infection (Nature 2011, DOI: 10.1038/nature10154). None of the inhibitors is yet in clinical trials, but the biotech firm ADRx has licensed the inhibitor-design method.
The 2005 work “was a breakthrough for amyloid structure studies and for crystallographic methodology,” says Robert Tycko, an amyloid expert at the National Institutes of Health. “So far, cross-β crystal structures of the full-length peptides and proteins have not been described, possibly because the inherent tendency of β-sheets to twist interferes with crystallization.” Studies of full-length fibril-forming proteins have been carried out with solid-state nuclear magnetic resonance spectroscopy instead.
“Crystallographers are notoriously successful in eventually crystallizing almost anything,” Tycko says. “So one should not rule out the possibility that cross-β structures of full-length amyloid-forming polypeptides will appear in the near future.”
C&EN's YEAR IN REVIEW
Top Headlines of 2015
- Chemical Makers Looked To Big Deals
- NASA Got Up Close And Personal With Pluto
- Opposition To Neonicotinoids Intensified
- 2015 Nobel Prizes In Science At A Glance
- Climate Pact Clinched
- Little Good News For Chemistry Job Outlook In 2015
- Pfizer To Merge Again, This Time With Allergan
- Oil And Gas Industry Under Pressure
- Greening Up Fracking
- An Industry In Spin Cycle
- Finally, Emoji For Chemists
- A Big Deal For Chemists
- Tianjin Explosion Put Spotlight On Safety
- Women Assumed Leadership Roles At American Chemical Society
- Artificial Ingredients In The Crosshairs
- Jacqueline K. Barton, Unwavering Chemistry Champion
- House Science Committee Chair Pummeled Science Agencies
- NIST Veteran Became U.S. Government's Top Chemist
- Gene-Editing Technique Raised Ethics Questions
- World Chemical Production At A Glance
- Classroom Fires During Science Demonstrations Spark Concern
- Climate Was Right For Deal-Making
- American Chemical Society Dives Deeper Into Open Access With The Debut Of ACS Omega
- Lego Began Research On Switching To A Biobased Plastic
- New Chair Took The Helm At Troubled Chemical Safety Board
- Genetically Modified Foods In The Spotlight
- Overhaul Of U.S. Chemical Law Moved
- ACS Scholars Program Turned 20
- American Chemical Society Expanded Its Global Reach
- Scientists Called For Standardized Antibodies
Top Research of 2015
- Flexible Electronics You Can Inject
- Special Delivery For Sensitive Reagents
- Yeast Programmed For Opioid Total Synthesis
- Miracle Machine Builds Molecules On Demand
- Nickel Shines As A Catalyst
- 3-D Printing Takes On A New Dimension
- Atomically Thin Films Grow In Number
- Digging In The Dirt Yields Novel Bacteria Fighter
- Keeping GMOs On A Leash
- A Liquid With Holes In It
- Electron Microscopy Provides Unprecedented Close-ups
Revisiting Research of 2005


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter